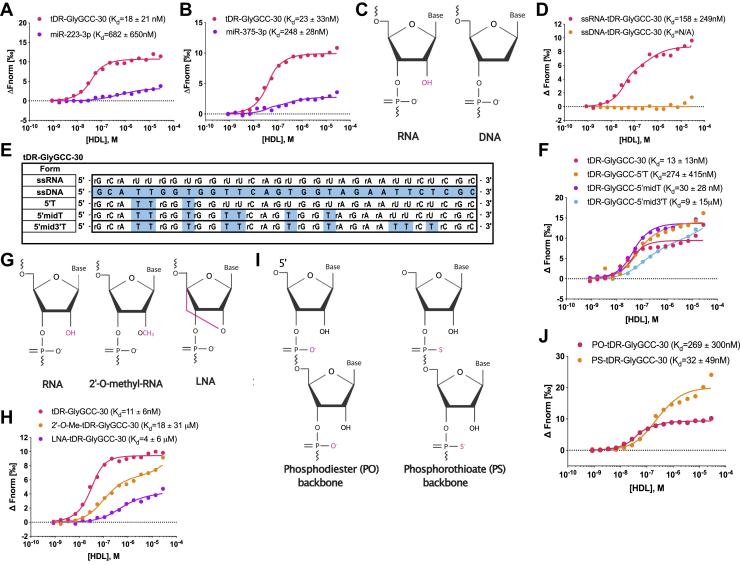
Figure 4.
Biochemical properties of tDR-GlyGCC binding to HDL using MST. MST analysis of interactions between DGUC-HDL and (A) miR-223-3p or (B) miR-375-3p. C, structure of ribonucleotides (RNA) and deoxyribonucleotides (DNA). Hydroxyl group on RNA highlighted. D, representative MST binding curves of single-stranded RNA (ssRNA) or single-stranded DNA (ssDNA) of tDR-GlyGCC-30. E, synthesized tDR-GlyGCC-30 sequences of ssRNA, ssDNA, and rU>T mutations: at the 5′ terminal end (5′T), the 5′ terminal end and middle (5′midT), and the entire oligonucleotide (5′mid3′T). F, MST binding curves of DGUC-HDL with tDR-GlyGCC-30 and its rU>T mutations from (E). G, structure of RNA with 2′-O-methylation (2′-O-Me) or locked-nucleic acid (LNA) which harbors a bridge between 2′ and 5′ positions of the base ring. Modifications highlighted. H, MST binding curves of DGUC-HDL and tDR-GlyGCC-30 with either 2′-O-Me or LNA modifications. I, structure of RNA with phosphodiester (PO) or phosphorothioate (PS) backbones. J, MST binding curves of DGUC-HDL and tDR-GlyGCC-30 with either PO or PS backbones. Binding affinities (Kd) of mean MST analysis from triplicate assays are presented, Kd are mean ± SD. Structures created with BioRender.com. DGUC, density-gradient ultracentrifugation; HDL, high-density lipoprotein; MST, microscale thermophoresis; tDR, tRNA-derived sRNA.