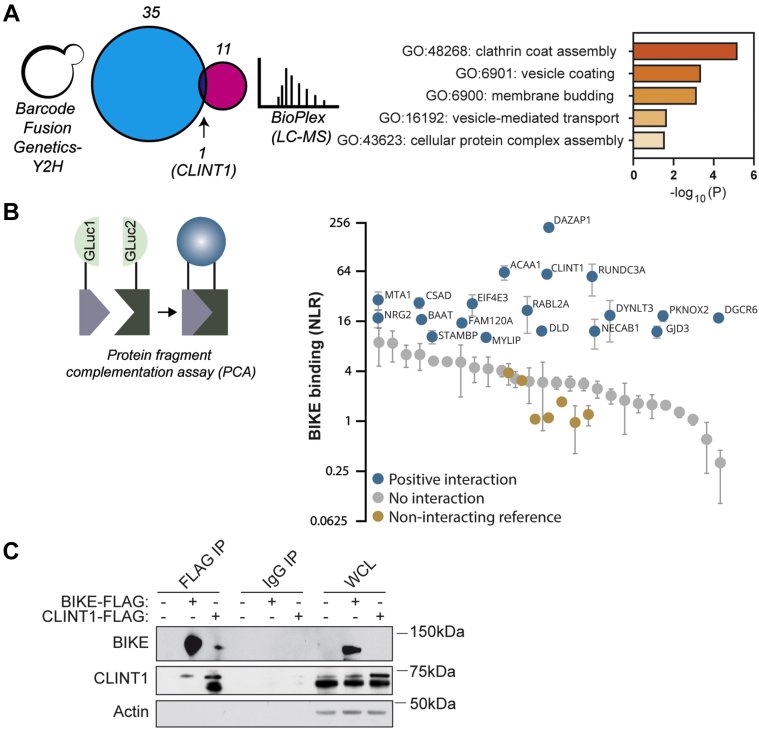
Figure 1.
High-throughput screen identifies BIKE interactors. A, left: Barcode Fusion Genetics-Yeast-2 Hybrid (BFG-Y2H) assay was carried out using barcoded baits and preys from the human ORFeome library that were physically linked with a split Cre recombinase following pooling of baits and preys postselection. Hits were defined by stringent cutoffs for the stickiness index and number of observations during sequencing. The Venn diagram shows the number of unique and overlapping hits emerging via the BFG-Y2H screen and the publicly available BioPlex dataset generated via an affinity purification MS approach. Right: Molecular function terms and p values derived from Gene Ontology (GO) enrichment analysis of BIKE, CLINT1, and their overlapping interactors following integration of the BIKE interactome data with BioGRID data via BiNGO. B, left: Schematic of the protein-fragment complementation assay (PCA) format. Right: BIKE interactions with 45 of the 47 putative interactors measured via PCAs in 293T cells. Dots depict the mean normalized luminescence ratio (NLR) and standard deviation (SD) of a representative experiment of 2 conducted with 3 replicates each. Gold, a reference set of 7 proteins that do not interact with BIKE. Gray, noninteracting proteins among the 47 putative BIKE interactors. Blue, BIKE-interacting proteins as defined by a cutoff of NLR>10 representing greater than two SDs above the mean NLR of the reference set. C, IPs with anti-FLAG antibody or IgG control from lysates of Huh7 cells ectopically expressing BIKE-FLAG, CLINT1-FLAG, or Empty-FLAG. Molecular weight markers are indicated on the right (kDa). Membranes were blotted with antibodies against BIKE, CLINT1, and actin. BIKE, BMP2-inducible kinase; CLINT1, clathrin-interacting protein 1; IP, immunoprecipitation; WCL, whole-cell lysate.