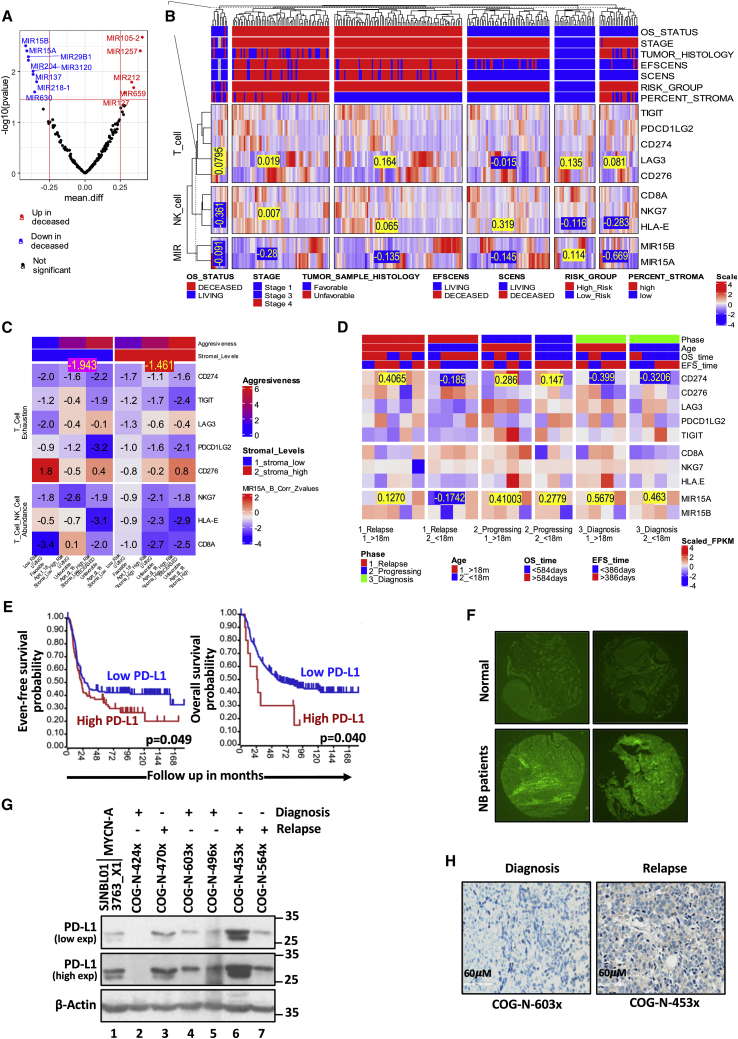
Figure 1.
High-risk NB patients and PDX tumors show higher tumor-infiltrating lymphocyte exhaustion signatures together with reduced miR-15a and miR-15b
(A) The Volcano plot depicting top-ranked up- (red) and down-regulated (blue) miRs (miR genes) in deceased versus living cohorts. The log-transformed TARGET dataset (n=249) was analyzed to prioritize miRs with statistically significant fold changes. The vertical red lines depict the fold change threshold of 1.77, corresponding to the log 2-fold change of 0.25. The horizontal red line indicates the P-value threshold of 0.03. The study prioritized miR-15a and miR-15b as tumor suppressor miRs for further characterization and validation. (B) The heatmap depicting the differential expression of miR-15a, miR-15b, and the T- and NK- cell signature genes across the TARGET dataset patient cohorts. The 249 TARGET dataset samples were classified into six categories with different aggressiveness based on a multi-attribute criterion depicted in the top annotation. The mean miR-15a and miR-15b log-transformed & scaled expression values for stromal-high and stromal-less tumor categories were -0.28 & -0.669. Two of the six categories were significantly enriched with stromal-high tumors. The values in stromal-high tumors were significantly lower than the corresponding mean values across the four other categories. This indicates potential antitumor immune response functions of miR-15a, and miR-15b. (C) Heat map of Pearson correlation Z-values between the mean miR-15a & miR-15b expression values and the eight genes of T- and NK- cell signatures. CD274 showed a strong signal with significant negative correlation profiles in stromal-low tumors versus the stromal-high tumors. This is consistent with the previous observations that miR-15a, miR-15b are tumor suppressors whose expression is higher in stromal-low tumors leading to better mR inhibitory functions captured as the significant negative correlations with target mRNA in these cohorts. (D) Heatmap depicting the differential expression of miR-15a, miR-15b, and the T- and NK- cell signature genes across the PDX dataset patient cohorts. The 24 PDX dataset samples were classified into six categories with different aggressiveness based on the Phase and Age depicted in the heatmap’s top annotation. The mean miR-15a scaled expression values for the two less aggressive categories were high (0.5679 & 0.463), while its target mRNA CD274 demonstrated suppressed expression (-0.399 & -0.3206), corroborating our hypotheses that miR-15a/miR-15b-CD274 axis might contribute to immune evasion and tumor aggressiveness. (E) Kaplan-Meier curves showing event-free and overall survival probability rates with different levels of PD-L1 in NB patients. (F) Immunofluorescence images of PD-L1 on NB patient tumor and normal tissue microarrays photographed at 5X. (G) Western blotting analysis of PD-L1 in whole lysates of PDX tumors of NB patients collected at the diagnosis and progression stages. Images were captured at low and higher exposures (exp). (H) IHC staining of PD-L1 in PDX tumors of NB patients at the diagnosis and progression stages.