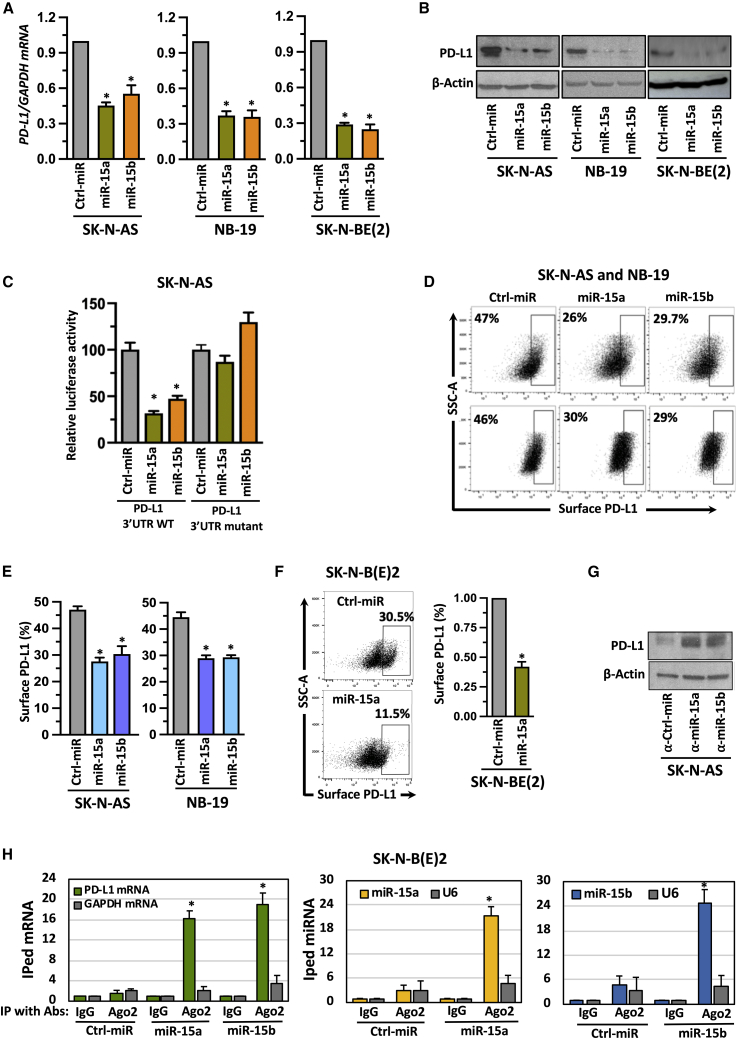
Figure 2.
MiR-15a and miR-15b target PD-L1 in NB cells
(A,B) A RT-qPCR quantification graph for PD-L1 mRNA (A), western blotting for PD-L1 total protein (B) in NB cells transfected with miR-15a, miR-15b, or control (ctrl) miRs for 48 h. (C) Luciferase reporter assay in SK-N-AS cells cotransfected with either luciferase reporter vector containing the PD-L1-3’UTR wild-type or mutant in the presence or absence of miR-15a and miR-15b for 48 h, followed by measuring the luciferase activity. (D-F) Representative flow cytometric plots for PD-L1 surface expression analyzed by using anti-PD-L1 phycoerythrin (PE) conjugated antibody in SK-N-AS and NB-19 cells transfected with miR-15a, miR-15b or ctrl miRs for 48 h (D and E) or miR-15a expressing stable SK-N-B(E)2 cells (F). (G) Western blotting for PD-L1 total protein in NB cells transfected with inhibitors of miRs such as α-miR-15a, α-miR-15b or α- ctrl miRs for 48 h. (H) A RT-qPCR quantification graph for Ago2-occupied PD-L1 mRNA (first panel), Ago2-occupied miR-15a (second panel), and Ago2-occupied miR-15b (third panel). SK-N-BE(2) cells were transfected with miR-15a, miR-15b, or ctrl miRs for 48 h followed by immunoprecipitation (IP) with α-Ago2 antibody. Ago2 bound RNA complexes were eluted, purified, and quantified for Ago2 bound mRNA and miRs by RT-qPCR using TaqMan assays. GAPDH mRNA and U6, a non-coding small nuclear RNA, were used as non-specific controls. The data were compared with the control IgG-bound mRNA or miR and set to one for normalization. Data represent the mean ± standard error of three to four independent biological experiments. Statistical analyses were performed using a two-sided unpaired t-test. *p < 0.001.