Figure 3.
eEF1A2 is a Cpmer-binding protein and similarly regulates CM differentiation
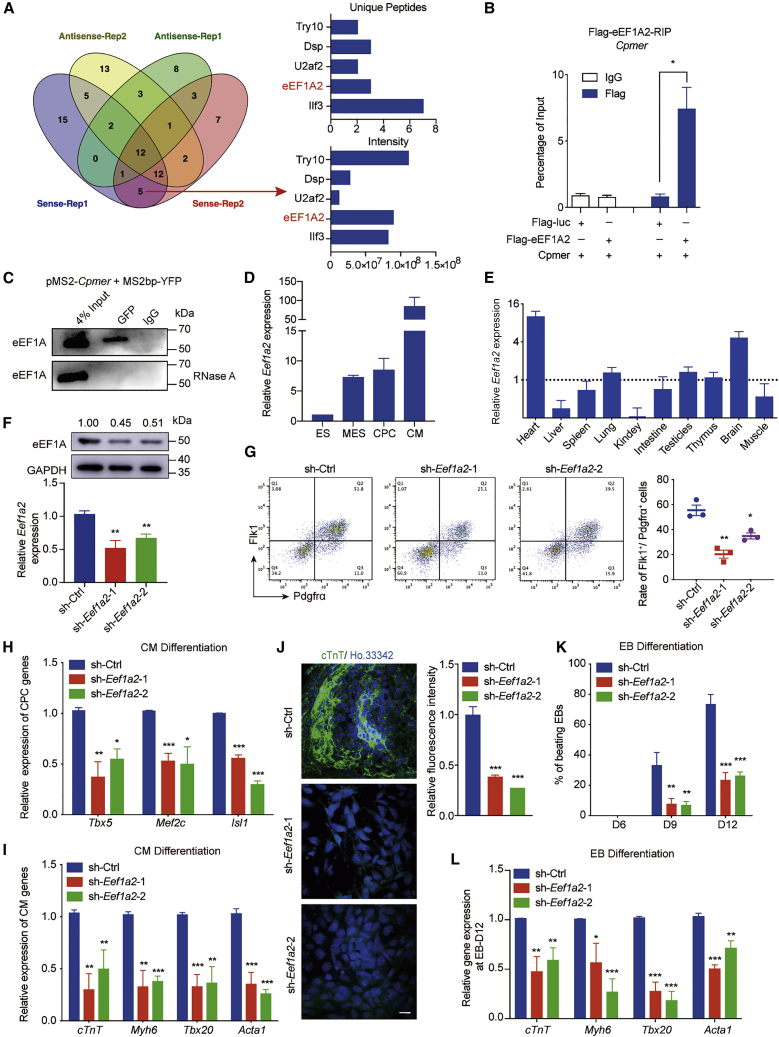
(A) MS to identify the potential Cpmer-binding proteins (left). The unique peptides and intensities of 5 candidate proteins are shown (right).
(B) RIP analysis for exogenous expression of Cpmer and FLAG-eEF1A2 in 293T cells, determined by an anti-FLAG antibody. Data shown are the mean ± SEM, n = 3 independent experiments.
(C) MS2bp-YFP RNA pull-down analysis for the co-transfected pMS2-Cpmer and MS2bp-YFP in MESs with or without RNase A treatment. An anti-GFP antibody was used to recognize the YFP protein.
(D) The expression pattern of Eef1a2. Data shown are the mean ± SEM, n = 3 independent experiments.
(E) The relative expression levels of Eef1a2 in neonatal mouse tissues. Data shown are the mean ± SEM, n = 3 mice.
(F) Detection of mRNA and protein expression levels of Eef1a2 at the MES stage after Eef1a2 knockdown. Data shown are the mean ± SEM, n = 3 independent experiments.
(G) FACS analysis and statistics of the percentage of Flk1+Pdgfrα+ cells. Data shown are the mean ± SEM, n = 3 independent experiments.
(H and I) qRT-PCR analysis of CPC (H) and CM (I) marker genes. Data shown are the mean ± SEM, n = 3 independent experiments.
(J) cTnT immunostaining (green) at the CM stage (left) and statistics for relative fluorescence intensity (right). Scale bar, 20 μm. Data shown are the mean ± SEM, n = 6 fields from 3 independent experiments.
(K) Percentage of spontaneously contracting EBs of EB differentiation. Data shown are the mean ± SEM, n = 3 independent experiments.
(L) qRT-PCR analysis of the CM marker genes. Data shown are the mean ± SEM, n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (versus sh-Ctrl); Student’s t test.
See also Table S2.