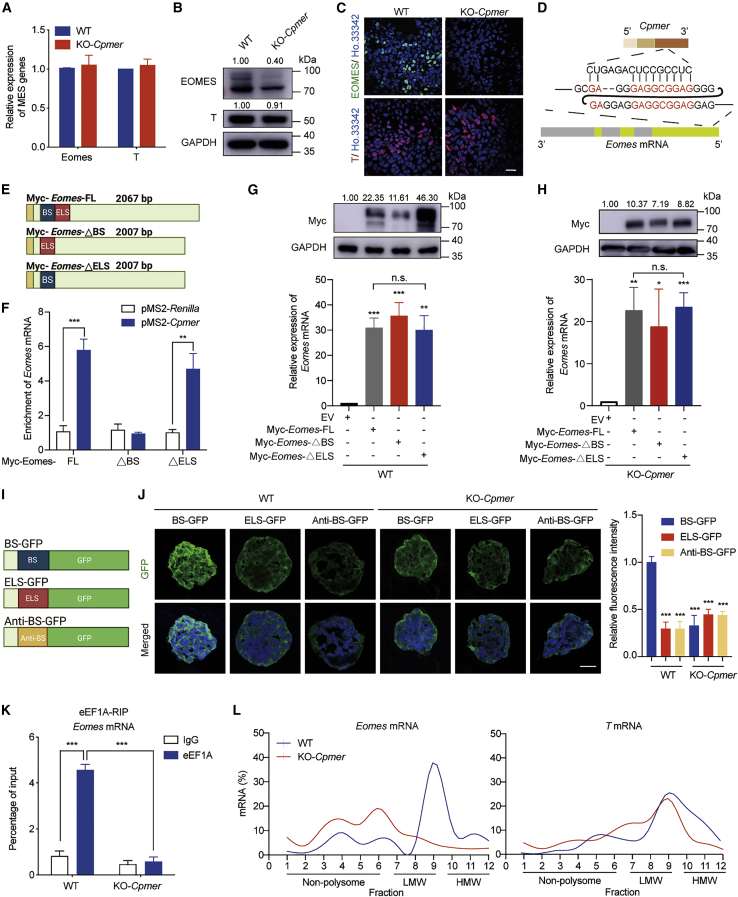
Figure 4.
Cpmer/eEF1A2 recognizes mRNA of the mesoderm gene Eomes and regulates its translation
(A–C) qRT-PCR (A), western blot (B), and immunofluorescence (C) detection of the expression of representative mesodermal marker genes (Eomes and T). Data shown are the mean ± SEM, n = 3 independent experiments. Scale bar, 20 μm.
(D) Regions of potential recognition between Cpmer and mouse Eomes mRNA.
(E) Schematic of the FL and deletion mutants of Eomes mRNA. BS, blue; ELS, red; Myc tag, yellow.
(F) MS2bp-YFP RNA pull-down analysis for exogenous expression of Cpmer with FL or deletion mutant Eomes mRNA in 293T cells. pMS2-Renilla RNA was used as a negative control. The fold enrichment relative to the immunoglobulin G (IgG) control is shown. Data shown are the mean ± SEM, n = 3 independent experiments.
(G and H) qRT-PCR and western blot analysis of Eomes mRNA and protein levels in WT (G) or KO-Cpmer (H) cells transfected with FL or deletion mutant Eomes. Data shown are the mean ± SEM, n = 3 independent experiments.
(I) Schematic of the GFP reporters. BS, blue; ELS, red; anti-BS, yellow.
(J) GFP signal analysis at EB-day 4 in cells transfected with distinct GFP reporters (left) and statistics for relative GFP signal intensity (right). Scale bar, 20 μm. Data shown are the mean ± SEM (n = 6 fields from 3 independent experiments).
(K) RIP analysis of the interaction of eEF1A with Eomes mRNA. Data shown are the mean ± SEM, n = 3 independent experiments.
(L) Percentage of Eomes and T mRNAs in the gradient total RNA, as measured by qRT-PCR in each fraction collected from the polysome profiling assay. LMW, low molecular weight; HMW, high molecular weight. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; n.s., not significant; Student’s t test.
See also Figures S3 and S4 and Table S3.