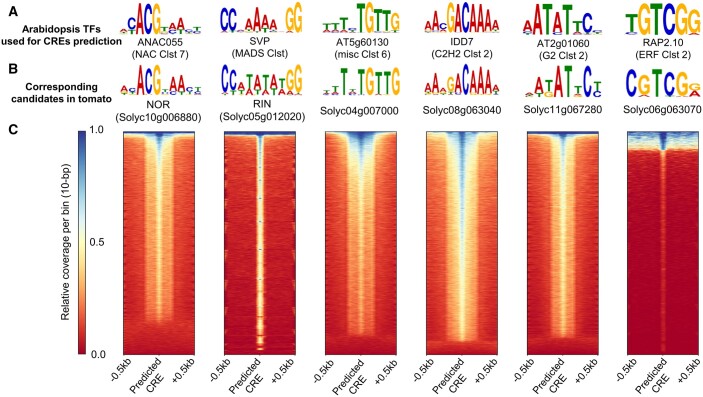
Figure 5.
Consistency between the DL-predicting CREs and the binding sites of tomato TFs. We selected six tomato TFs (NOR, RIN, Solyc04g007000, Solyc08g063040, Solyc11g067280, and Solyc06g063070), which were the orthologs of the genes in NAC Clst 7, MADS Clst, misc Clst 6, C2H2 Clst 2, G2 Clst 2, and ERF Clst 2, respectively. A, Representative enriched motifs in the Arabidopsis DAP-Seq peaks (O’Malley et al., 2016) for the six TFs with the highest cumulative relevance to the genes significantly upregulated in the BR stage (see Figure 3E). B, The most probable enriched motifs in the DAP-Seq peaks for the described six tomato TFs, which exhibited similar sequence patterns to the corresponding Arabidopsis orthologs. C, Heatmaps for the relative read coverages surrounding the CREs predicted by each DL model. For all of the six TFs, most predicted CREs were enriched with DAP-seq reads, indicating TF binding.