Figure 6.
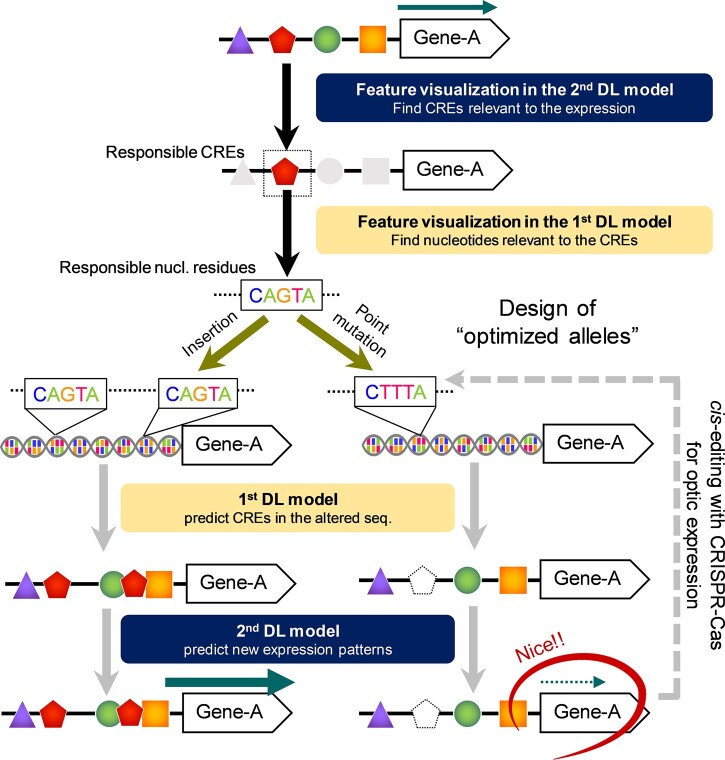
Model for expression design based on explainable DL. If the objective expression patterns can be well predicted from CRE arrays, two-step feature visualization in the prediction models (or the second and then first DL models, see Figure 1B) will allow identification of the nucleotide-scale factor(s) responsible for the expression pattern. Randomization of the responsible residues can derive potentially unlimited variations for the objective expression pattern, which can be easily predicted using the first and second DL models. Once a desirable expression pattern is predicted, cis-editing with the CRISPR–Cas system may realize the design of the optimized allele.