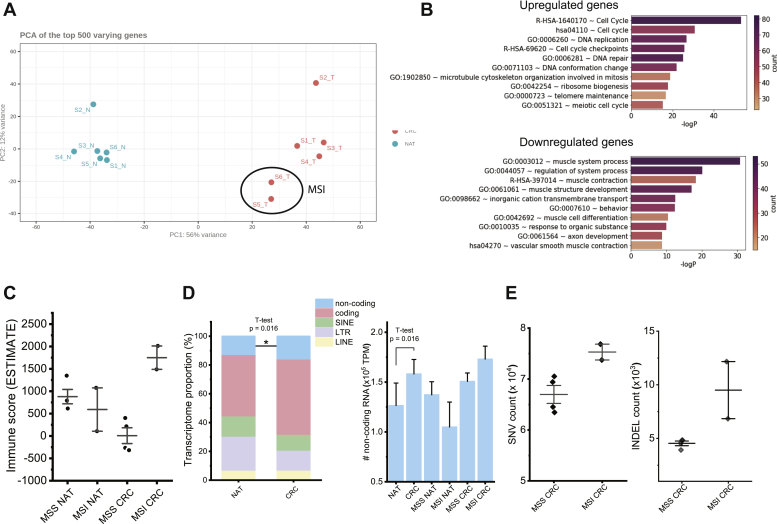
Fig. 2.
Transcriptomic profile of primary tumor/normal adjacent tissue CRC biopsies. A, principal component analysis (PCA) of the top 500 varying genes of each tumor/NAT sample following paired-end RNA-Seq and gene read count normalization with DESeq2. MSI tissues (as determined by MSISensor) are encircled. B, GO term analysis of genes up/downregulated in CRC tissues compared with their adjacent NAT. Genes submitted to GO term analysis were those with ∣log2FC∣ >1 and that were found to be differentially regulated in all samples, using TPM normalized values. C, bar graph showing the mean ESTIMATE immune score of MSS NAT, MSI NAT, MSS CRC, and MSI CRC, with standard deviation shown. D, stacked bar graph showing the mean proportion of the transcriptome attributable to five distinct transcript biotypes in NAT versus CRC samples, with the differences in the proportion of noncoding transcripts being statistically significant between NAT and CRC (noncoding: p = 0.016; coding: p = 0.078; SINE: p = 0.15; LTR: p = 0.056; LINE: p = 0.95). E, scatterplots displaying the SNV counts and INDEL counts of MSS and MSI CRC tissues determined by SNPEff genomic annotation, with mean and standard error bars. CRC, colorectal cancer; MSI, microsatellite instability; MSS, microsatellite stable; NAT, normal adjacent tissue; TPM; transcripts per million.