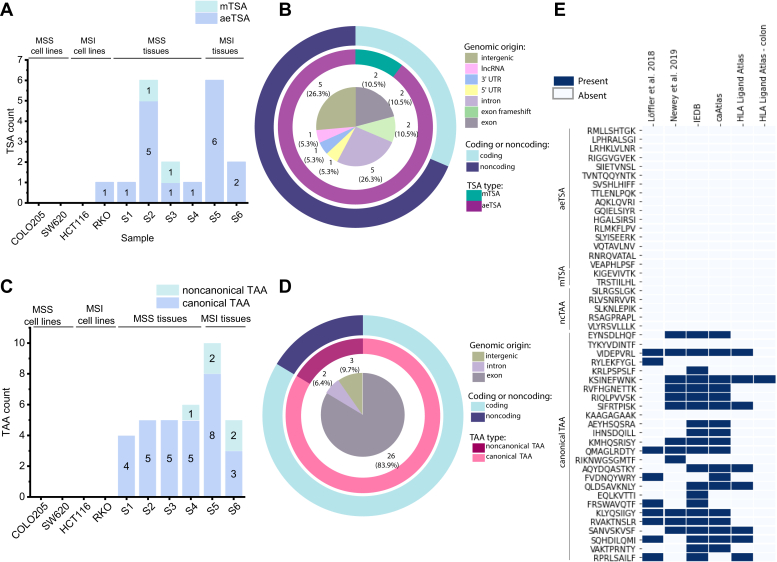
Fig. 4.
Novel TSAs identified in colorectal cancer derive primarily from noncoding regions, whereas the majority of TAAs derive from exons. A, bar chart displaying the number of TSAs identified per sample. B, stacked pie chart identifying the genomic origin of TSAs in the inner pie, as well as what proportion of TSAs are mutated in the middle pie. The outer pie demonstrates what proportion of TSAs are from coding or noncoding sequences. C, bar chart displaying the number of TAAs identified per sample. D, stacked pie chart identifying the genomic origin of TAAs in the inner pie, and what proportion of TAAs are canonical or noncanonical in the middle pie. The outer pie displays what proportion of TAAs are from coding or noncoding sequences. E, heatmap displaying the presence or absence of putative TSAs and TAAs in two previous publications on CRC immunopeptidomics (Löffler et al. 2018 and Newey et al. 2019), as well as IEDB and HLA Ligand Atlas (all tissues, and only colon tissue). MSI, microsatellite instability; MSS, microsatellite stable; TAA, tumor-associated antigen; TSA, tumor-specific antigen.