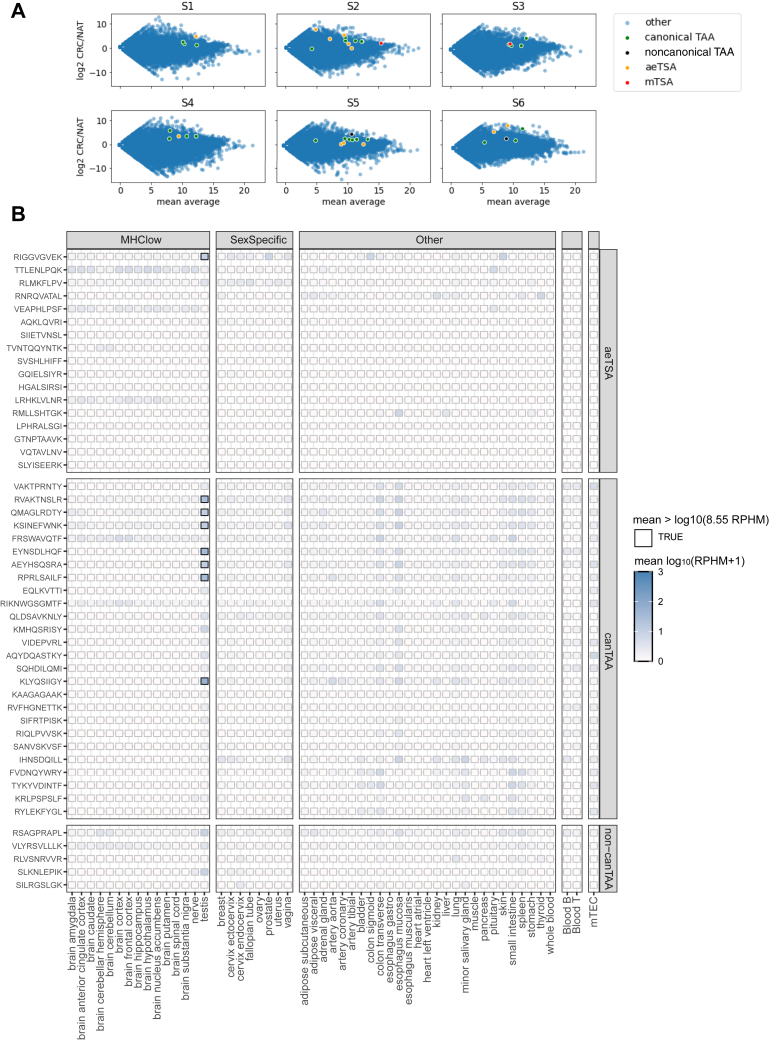
Fig. 5.
RNA expression profiles of putative TSAs and TAAs. A, scatter plots displaying the log2FC of transcripts, in TPM, in CRC compared with the matched NAT on the y-axis and the mean average expression in a given tissue sample (mean of CRC and NAT). Highlighted points indicate the source transcripts of putative TAA and TSAs. Both S4 and S5 plots have a canonical TAA point that is not visible, as it overlaps with another canonical TAA source transcript. B, heatmap of mean RNA expression in log(rphm+1) of aeTSA coding sequences and TAA coding sequences (divided as canonical TAAs [canTAA] and noncanonical TAAs [non-canTAA]) in normal tissues from Genotype Tissue Expression (GTEx) Portal and in pooled thymic epithelial cell samples. MHClow tissues include those from brain, nerve, and testis, which have been shown to lowly express MHC I. A black outline indicates a mean RNA expression >8.55 rphm. aeTSA, aberrantly expressed tumor-specific antigen; CRC, colorectal cancer; NAT, normal adjacent tissue; TAA, tumor-associated antigen; TSA, tumor-specific antigen.