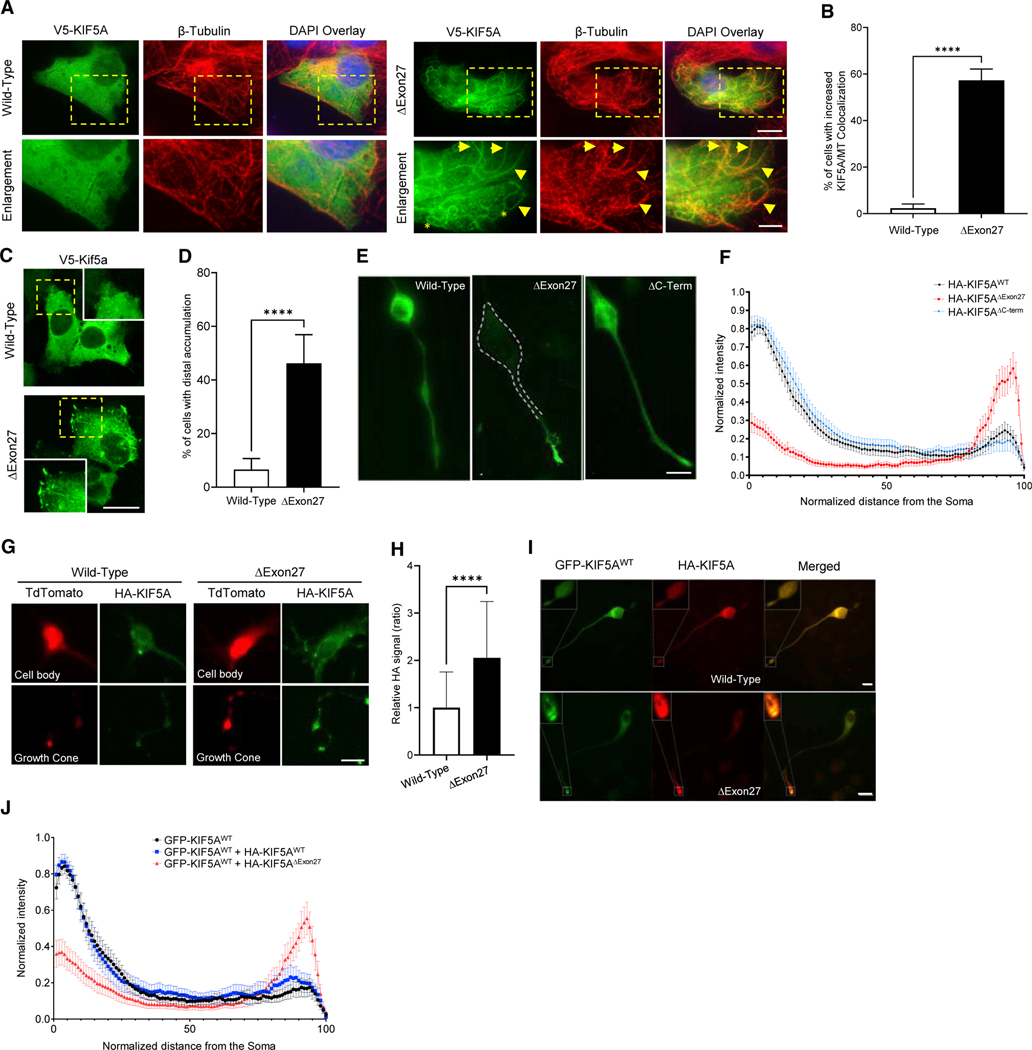
Figure 2. Mutant KIF5A associates more readily with microtubules, displays microtubule plus-end accumulation, and has a dominant-negative effect on wild-type KIF5A.
(A) SKNAS cells expressing V5-tagged KIF5AΔExon27 show increased microtubule (MT) co-localization compared with KIF5AWT as demonstrated by V5-KIF5A highlighting the MT tracks. Examples of KIF5A (V5; green) and β-tubulin (red) co-localization are indicated by arrowheads. Many cells have KIF5AΔExon27-associated MTs with a non-radial pattern (asterisks). Scale bars, 10 mm (wide view), 5 mm (enlargement).
(B) Quantification of the experiment in (A). n = 5 biological replicates are shown with p < 0.0001.
(C, E, and G). Expression of KIF5AΔExon27 results in distal/growth cone accumulation of tagged-KIF5A in transfected SKNAS (C), differentiated N2A (E), and PMN cells (G). Scale bars, 20 mm (C), 25 μm (E), and 10 μm (G).
(D) Quantification of the percentage of transfected SKNAS cells in (C) with distal accumulation. n = 5 biological replicates are shown with p < 0.0001.
(F) HA intensity analysis of the experiment in (E). n = 3 biological replicates are shown with n ≥ 60 cells per sample.
(H) Quantification of the HA signal intensity from the growth cone compared with that of the cell body for the PMNs in (G). n = 3 biological replicates are shown with p < 0.0001.
(I) Representative images of differentiated N2A cells transfected with GFP-KIF5AWT and either HA-tagged KIF5AWT or KIF5AΔExon27. Scale bar, 25 μm. Confirmation of this protein binding is shown in Figure S1B.
(J) Quantification of the GFP intensity along the length of the cells in (I) when different forms of HA-tagged KIF5A are present. n = 3 biological replicates with n≥51 cells analyzed per sample. Data in (B), (D), and (H) are represented as mean ± SD. Data in (F) and (J) are represented as mean ± 95% CI.