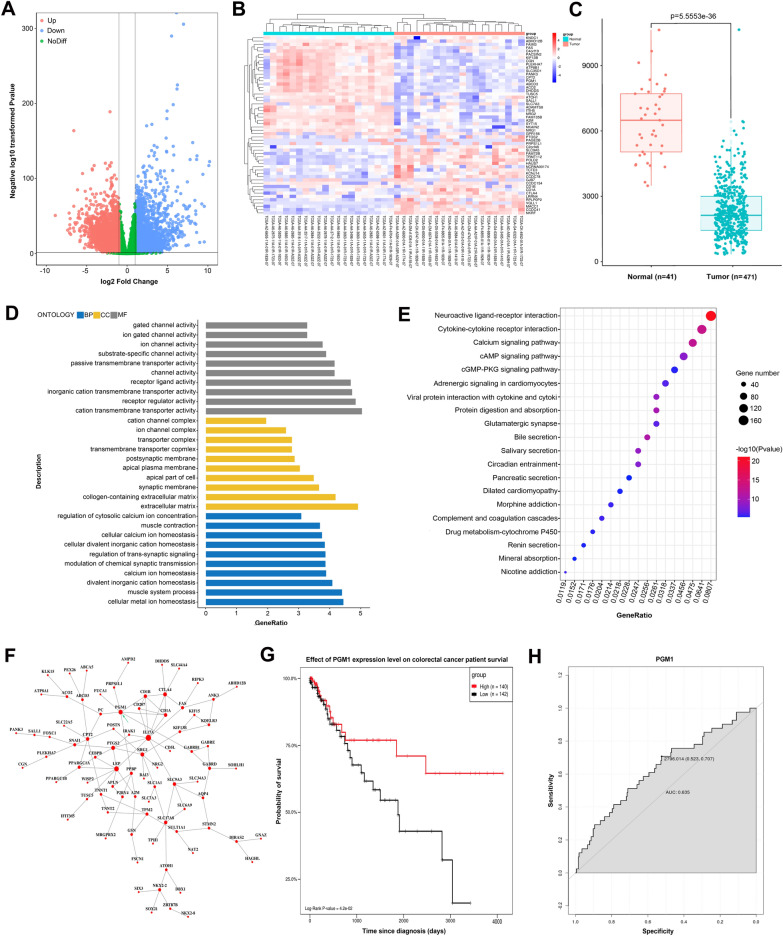
Fig. 1.
Identification of PGM1 via TCGA datasets. A Volcano plot showing the distribution of DEGs in TCGA. Red color shows up-regulated genes in normal tissue while blue color shows down-regulated genes in tumor tissue. B A heat map of 54 core genes from the TCGA-COAD dataset comparing CRC tissue and adjoining normal tissue. C PGM1 expression in normal control and CRC patients based on samples listed in the TCGA database (matched TCGA normal and GTEx data, P < 0.001). Each dot represents one sample; D GO network showing relationships between DEGs and predicted functions. Vertical axis, GO category; horizontal axis, P-values. A lower P-value indicates a greater predicted involvement of the DEG in CRC+. E KEGG network showing connections between pathways and DEGs. Vertical axis, pathway classification; horizontal axis, P-value. A lower P-value indicates greater numbers of pathways involving DEGs. F PPI (protein–protein interaction) network of DEGs as drawn by Cytoscape. G Kaplan–Meier analysis showing the association between PGM1 expression and overall survival among CRC patients. H AUC curves with respect to PGM1 gene expression in the TCGA cohort. PGM, phosphoglucomutase; CRC, colorectal cancer; TCGA, The Cancer Genome Atlas; AUC, area under the curve