Fig. 3. bGSDMs are activated by proteolytic cleavage.
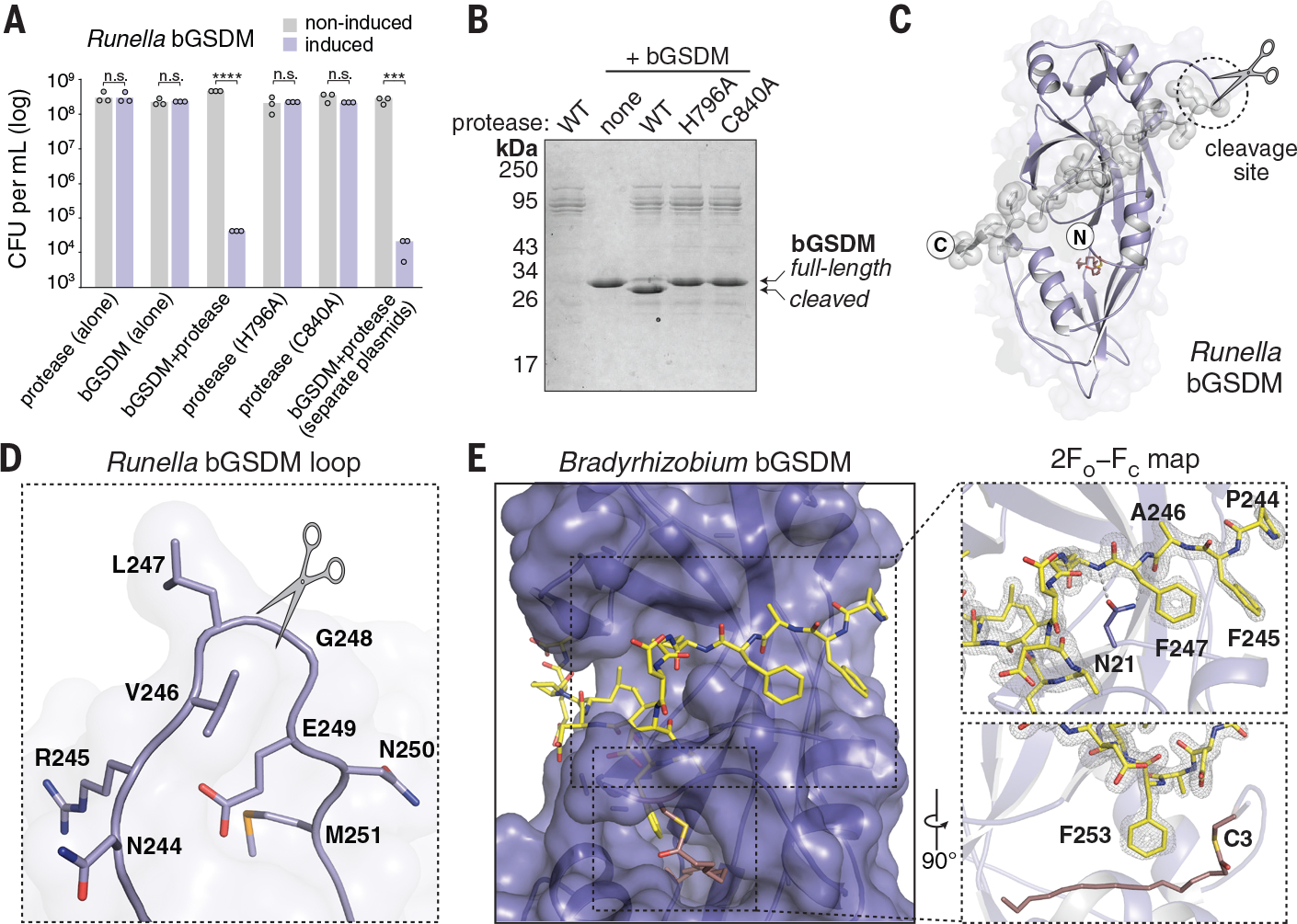
(A) Toxicity of Runella bGSDM in vivo requires the associated protease. Bacteria expressing WT and mutated versions of the Runella bGSDM–protease operon were grown on LB-agar in conditions that repress or induce expression. Data represent colony-forming units (CFU) per milliliter, and bar graphs represent an average of three independent replicates, with individual data points overlaid. Asterisks indicate statistically significant differences compared with the respective noninduced control using two-sided t-test. n.s. ≥ 0.05; ***P = 0.0001–0.001; ****P < 0.0001. (B) Runella bGSDM cleavage by its associated protease is dependent on catalytic histidine and residues in vitro. 15% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) were run after cleavage at room temperature for 18 h and visualized by Coomassie staining. (C) The Runella bGSDM crystal structure and protease cleavage site. The Runella bGSDM structure is shown in lavender with the last 21 amino acids highlighted as gray spheres. (D) Close-up view of the Runella bGSDM cleavage site wherein cleavage occurs after the P1 L247 residue. (E) Structural overview of the Bradyrhizobium CTD and autoinhibitory interactions. The bGSDM is colored purple except for its last 16 residues, which are colored yellow. Insets show interactions of F245 and F247 adjacent to D21 of the N-terminal β sheet (top) and F253 and the palmitoyl modification at C3 (bottom). The 2FO−FC (contoured at 1.5 σ) map is shown as gray mesh fit to the last 16 residues.
