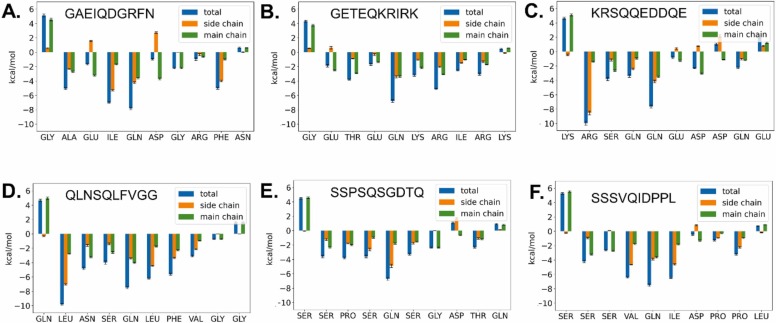
Fig. 2.
Per-residue binding energy decomposition of peptides. A) GAEIQDGRFN. B) GETEQKRIRK. C) KRSQQEDDQE. D) QLNSQLFVGG. E) SSPSQSGDTQ. F) SSSVQIDPPL. Using MM/GBSA analysis, the interaction energy of each amino acid with the rest of the system was estimated. The importance of the residues at P1 and P2 positions become apparent. Variations at e.g. P4 and P1’ positions could also explain improved binding of the corresponding peptides. Values are given in kcal/mol.