Abstract
Background
Extracellular vesicles (EVs) are regulators of cell–cell interactions and mediators of horizontal transfer of bioactive molecules between cells. EV-mediated cell–cell interactions play roles in physiological and pathophysiological processes, which maybe modulated by exposure to pathogens and cocaine use. However, the effect of pathogens and cocaine use on EV composition and function are not fully understood.
Results
Here, we used systems biology and multi-omics analysis to show that HIV infection (HIV +) and cocaine (COC) use (COC +) promote the release of semen-derived EVs (SEV) with dysregulated extracellular proteome (exProtein), miRNAome (exmiR), and exmiR networks. Integrating SEV proteome and miRNAome revealed a significant decrease in the enrichment of disease-associated, brain-enriched, and HIV-associated miR-128-3p (miR-128) in HIV + COC + SEV with a concomitant increase in miR-128 targets—PEAK1 and RND3/RhoE. Using two-dimensional-substrate single cell haptotaxis, we observed that in the presence of HIV + COC + SEV, contact guidance provided by the extracellular matrix (ECM, collagen type 1) network facilitated far-ranging haptotactic cues that guided monocytes over longer distances. Functionalizing SEV with a miR-128 mimic revealed that the strategic changes in monocyte haptotaxis are in large part the result of SEV-associated miR-128.
Conclusions
We propose that compositionally and functionally distinct HIV + COC + and HIV–COC– SEVs and their exmiR networks may provide cells relevant but divergent haptotactic guidance in the absence of chemotactic cues, under both physiological and pathophysiological conditions.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-04068-2.
Keywords: Small RNA-Seq, Proteomics, Extracellular vesicles, Cocaine, miR-128-3p, Haptotaxis
Background
Extracellular (ex) molecules including exProteins and exRNA, such as miRNA and other non-coding RNAs are present in circulation and are implicated in various physiological and pathological processes. The levels of ex-molecules may be regulated by licit and illicit substance (such as cocaine), as well as pathogens, including bacteria, fungi, and viruses, such as the Human Immunodeficiency Virus (HIV). Proteomics analysis of human semen and blood showed that a good number of exProteins are associated with EVs [1], while others are not EV-associated. Moreover, it has been shown that different RNA biotypes are present in semen, some of which are SEV-associated [2, 3]. SEV have critical roles in a variety of biological processes, such as: regulation of HIV infection [4–11], inflammation [8, 12], transcription factors [6], and male fertility [13, 14]. Interestingly, proteomic studies have shown spectra of altered proteome between SEV and blood-derived EVs (BEV) [1], as well as HIV-induced changes in SEV proteome [1]. These prior studies suggest that the condition of the host, such as infection with HIV and psychostimulant use, may trigger significant alterations in EV composition and functions.
Psychostimulant use in HIV-infected individuals, including cocaine [15], contributes to rapid disease progression and virus-induced pathology [16–21]. In addition, drugs of abuse may affect male fertility [22], although some fertility parameters may not be altered in HIV-infected men unless they progressed to AIDS [23–25]. However, studies reporting on the pathogenesis of combined HIV-infection and cocaine use are limited [25–27]. Efforts to elucidate biomolecular signatures of disorders have been undertaken by analyzing body fluids, especially blood, with either proteomics or RNA-Seq approaches [28–35]. However, it is likely that neither approach is sufficient given the biological dynamics and complexity of body fluids and their extracellular molecules. For example, independent profiling of SEV proteome [1] and miRNAome [3, 36] show that SEV have complex molecular networks. Thus, systems biology and multi-omics approaches that integrate proteome and miRNAome networks may facilitate elucidation of the molecular signature of SEV associated with reproduction, health, and disease [37–39], along with their potential functions.
Of particular interest is the finding that SEV from healthy, HIV + COC + men alter monocyte actin cytoskeleton dynamics, morphology, adhesion, matrix metalloproteinases secretion, and chemotactic migration [40]. Biologically, monocytes play a vital role in host immune responses to pathogens, where their locomotion, including migration (regulated by the adhesion molecules, chemokines, and pathogen-associated pattern-recognition receptors) [41, 42] are essential for their functions [43]. While there are different types of cell locomotion, directional or ballistic locomotion (cell migration in one direction) is expected to move cells rapidly between points, while random locomotion enables cells to be more exploratory. In random locomotion, cells exhibit non-directional migration that may allow them to sample the environment in search of information. This form of cell locomotion lacks sensory cues. Ballistic locomotion, which has been described for immune cells, aims at increased target identification [44] and involves multiple factors that may include one or a combination of external guidance cues. Such cues are either chemical, an intrinsic propensity of cells to continue moving in the same direction (persistence) in the absence of external guiding factor, or physical (ECM networks, such as collagen fibers). At baseline, cells may adopt random locomotion. However, if presented with a stimulus or guidance cue, the nature of the cue determines the rate and type of directional locomotion. In the presence of soluble cues, cells undergo chemotaxis [45–48], while haptotaxis occurs in response to ECM-anchored guidance cues [49–51].
Indeed, monocyte migration into tissues (important in viral pathogenesis and inflammation) requires intricate interplay between the cell surface and the ECM, which serves as the guidance cue. Depending on the guidance cues in the environment, monocytes can transmigrate from peripheral blood into inflammatory tissues or reverse-transmigrate from tissues to peripheral blood. Although altered levels of various exProteins and exRNAs have been observed in SEV, how they are regulated by HIV and cocaine use are not fully understood. Also not known is the role of SEV or SEV-associated molecules in regulating haptotaxis.
In the present study, we used SEV to study how HIV infection and cocaine use regulate the repertoire and functions of EV-associated proteins, miRNA, and their interactome. After paired proteomics and small RNA-Sequencing (sRNA-Seq) analyses, we employed an in vitro two-dimensional-substrate single cell haptotaxis assay for quantitative assessment of the effect of different SEV on haptotaxis. Finally, we functionalized SEV with exogenous miR-128 to directly address the role of SEV-associated miR-128 on monocyte haptotaxis.
Results
Study design
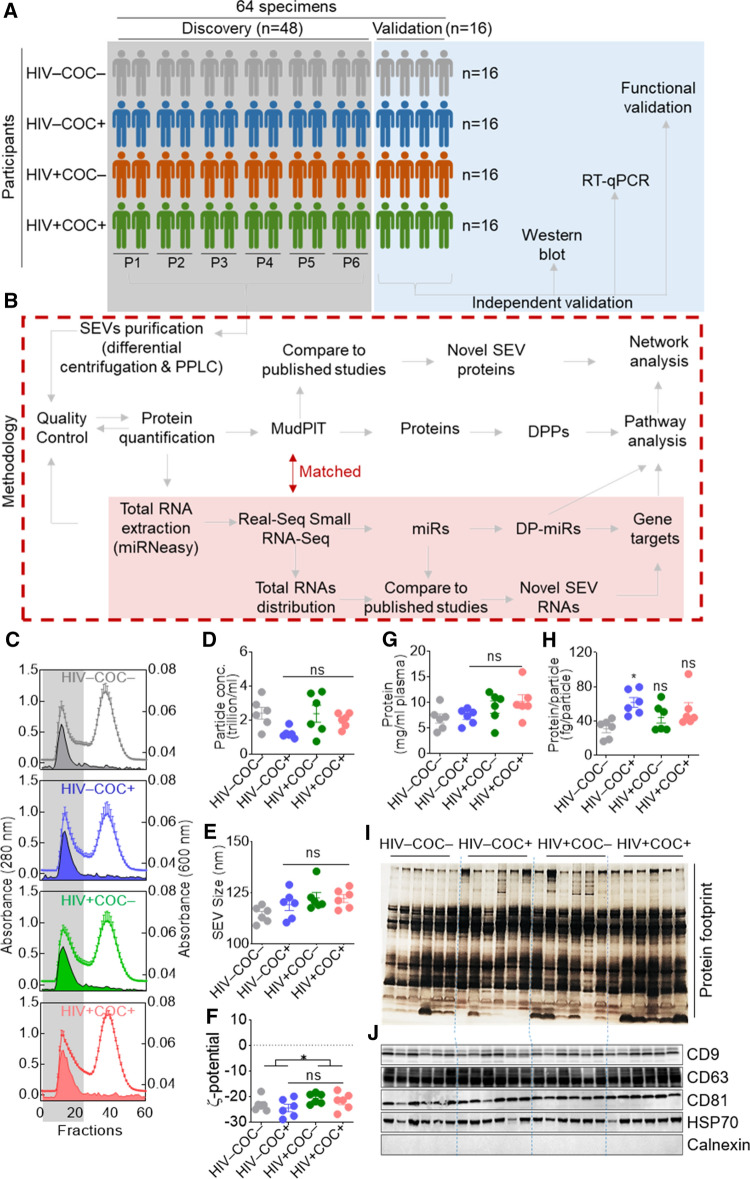
The cohort description, demographic information, and clinical characteristics of the study participants were previously reported [40]. Briefly, existing de-identified samples of human semen were obtained from participants (aged 18–81 years) in the Multicenter AIDS Cohort Study (MACS) [52, 53]. The MACS is a prospective cohort study of the natural history of HIV infection in homosexual and bisexual men, which was initiated in 1984 in 4 US sites (Baltimore-Washington DC, Chicago, Pittsburgh, and Los Angeles). Semen samples were obtained from study participants semi-annually from 1984 to 1987 and stored at − 80 °C until used. The participants included HIV uninfected (HIV −) and HIV-infected (HIV +) men who, at the time of semen collection, reported either using or not using cocaine. Participants were classified as cocaine users and included in the study only if they reported using cocaine (taken by any route). In other words, if participants reported using other all psychostimulants without cocaine, they were excluded [40]. HIV − participants had no history of HIV, hepatitis B virus (HBV), or hepatitis C virus (HCV) infections. HIV + donors were ART-naive, since samples were collected prior to the use of ART. SEV were isolated from 48 seminal plasma samples of HIV–COC– (n = 12), HIV–COC + (n = 12), HIV + COC– (n = 12) and HIV + COC + (n = 12) participants (Fig. 1A, gray shading) by differential centrifugation followed by a size exclusion chromatography purification, using particle purification liquid chromatography (PPLC) [2]. Isolated SEV were subjected to an integrative omics approach, consisting of proteomics analysis concurrent with an unbiased sRNA-Seq. For the discovery phase, a sample-pooling strategy (P1 to P6, Fig. 1A) that consists of pooling two samples into one biological replicate was used. Validation studies were conducted with SEV isolated from 16 independent donors (n = 4/group, Fig. 1A, blue shading). The processes of SEV isolation, omics, and bioinformatics analyses are summarized in (Fig. 1B, broken red line).
Fig. 1.
Study workflow and EV isolation. A Overall study design and sample description. B Methodological workflow. C Isolation spectra of SEV. Open histograms and filled areas represent absorbance at 280 nm and 600 nm, respectively. The longitudinal gray bar denotes EV-containing fractions. D Particle concentration, size, and protein concentration of SEV as indicated on the Y axis. E) Protein footprint by silver staining (top) and western blot of EV markers (bottom). F Zeta-potential (ζ-potential) of SEV. Error bars represent S.E.M. of 6 pools (n = 2) per group. Group comparison was achieved with an ordinary one-way Anova test (Tukey correction). *, Adj. p value < 0.05; ns, not significant
The physical properties of SEV from HIV − and HIV + participants who use or do not use cocaine
PPLC fractions encompassing the void peak, which contained SEV (Fig. 1C, gray shading) were collected, pooled, aliquoted, and stored at − 80 °C. The SEV were analyzed for particle concentration, which ranged between 0.74 and 3.66 trillion particles/mL of seminal plasma, with no significant differences among groups (Fig. 1D). A mean particle size of 114.3 nm, 119.1 nm, 122.5 nm, and 122.2 nm were measured for HIV–COC–, HIV–COC + , HIV + COC–, and HIV + COC + , respectively (Fig. 1E). Zeta-potential measurements ranged between − 19.6 and − 29 mV, indicating an overall negative surface charge of SEVs, with a decreasing trend of this negative charge upon HIV infection (Fig. 1F). This is in line with our previous findings [40], but in disagreement with another study [54]. The discrepancy between the studies may be due to the effect of ART that appears to oppose the decreased negative charge caused by HIV infection [54]. The protein concentration was also variable and ranged between 4.14 and 15.9 mg/mL of seminal plasma, with a non-significant increasing trend of the average concentration of 6.86, 7.28, 9.09, and 10.17 mg/mL, for HIV–COC–, HIV–COC + , HIV + COC–, and HIV + COC + , respectively (Fig. 1G). Interestingly, this increasing concentration trend remained after normalization of protein concentration to particle concentration, despite the high variance amongst samples with a protein weight per vesicle ranging between 16 and 95 fg/particle (Fig. 1H). This analysis was corroborated by a silver stain protein profiling which pointed to donor-dependent variations in the protein footprint and enrichment (Fig. 1I). All samples were positive for EV-markers—tetraspanins (CD63, CD9, CD81) and HSP70, and negative for calnexin, an endoplasmic reticulum marker used as negative control (Fig. 1J). Of note, the SEV samples used in this study had been characterized in a prior study, where morphological analysis by transmission electron microscopy (TEM, wide-field and close-up images), HIV reverse transcriptase activity, as well as the amount of HIV p24 and the levels of cocaine metabolite—benzoylecgonine were reported [40]. Together, these results indicated that the vesicles are EVs, and that the isolation was successful.
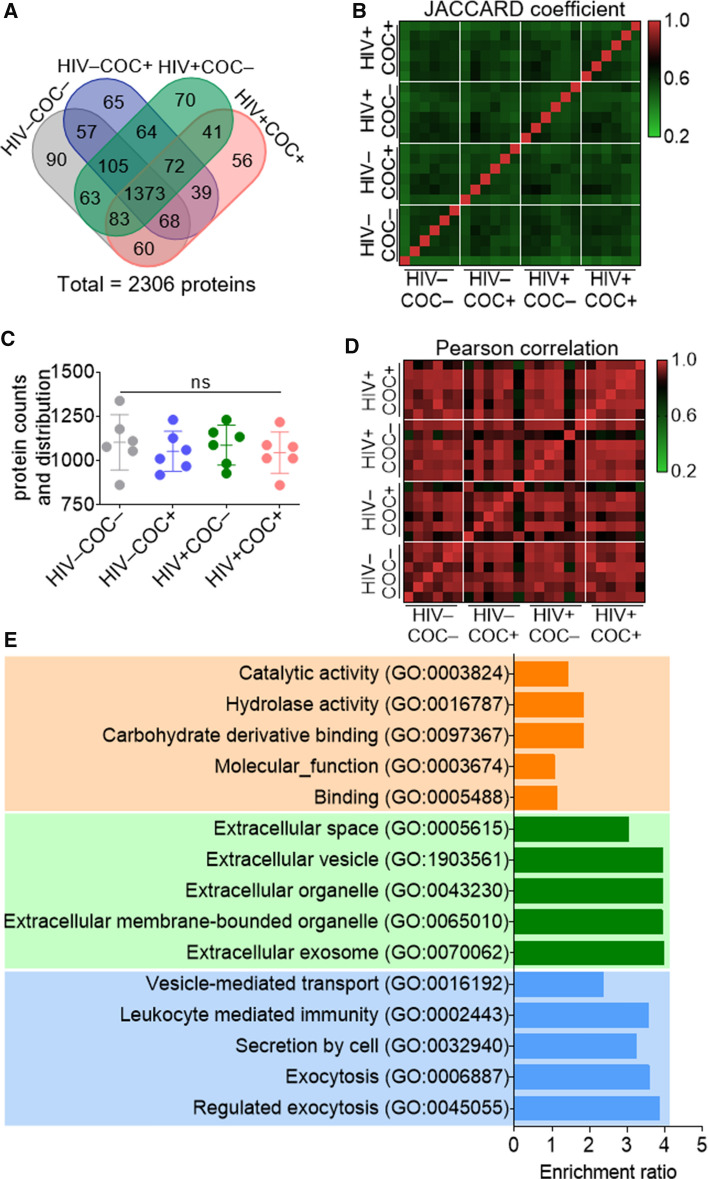
Characterizing the SEV proteome
Multidimensional protein identification technology—MudPIT [55] was used to analyze HIV–COC–, HIV–COC + , HIV + COC–, and HIV + COC + SEV proteomes. After mapping the peak lists to an in-house constructed database, the analysis yielded a total of 2306 proteins, of which 442 proteins (19%) were identified with a single unique peptide (Table S1). Protein distribution was balanced among the samples with no apparent differences between the groups (Figs. 2A, C). Intra- and inter-group variations were remarkably low as depicted by the JACCARD coefficient heatmap (Fig. 2B), a quantitative measure of the degree of similarity between various categories [56], and the Pearson correlation analysis (Fig. 2D). To contextualize our findings, this proteomics data set was compared to previously published SEV proteomes [57–60]. To this end a systematic literature search was conducted (Fig. S1A). The identified proteomics studies differed in their cohorts, SEV isolation methods, criteria for peptide selection, and/or output protein IDs (Table S2). The data sets were thus unified by removing redundant IDs, converting IDs to gene symbols using David gene ID conversion tool, and extracting IDs that are mappable to WEB-based GEne SeT AnaLysis Toolkit (See Supplemental File). A comparative Venn analysis of the remaining IDs revealed that SEV proteome is composed of at least 4796 SEV proteins, of which 858 are novel to the present study (Fig. S1B). Comparing the SEV proteome to Exocarta and Vesiclepedia EV databases, we found that 23.6% (1132 proteins) were previously not reported in EVs (Fig. S1C). Of the 858 novel SEV proteins identified in this study, 358 proteins have never been reported in any other EV studies and thus, are also novel EV proteins (Fig. S1D). Global GO-Term analysis of the total SEV proteome (2306 proteins) show enrichment in leukocyte mediated immunity among the Top-5 terms in biological processes, in addition to the exocytosis related terms (Fig. 2E). Cellular components were mostly involved in extracellular space terms, as expected, whereas molecular function was enriched in terms related to catalytic activity, hydrolase activity, carbohydrate derivative binding, and binding (Fig. 2E). A deeper analysis of the other significantly enriched GO biological processes reveal two functional themes of interest: the first relates to cell migration, cell motility, cell movement and chemotaxis, and the second pertains to immune activation, mainly involving leukocytes (Table S3). These GO Terms are in line with previously described SEV-related GO Terms as well as SEV-regulated functions [40].
Fig. 2.
Global analysis of the SEV Proteome from healthy or HIV infected participants who use or do not use cocaine. A Venn diagram showing common and unique proteins among the clinical groups. B Heatmap of the Jaccard coefficient depicting similarities and differences among clinical groups. C Protein counts and distribution among the clinical groups. Error bars represent S.E.M. of 6 pools (n = 2) per group. Group comparison was achieved with an ordinary one-way Anova test (Tukey correction); ns, not significant. D Pearson correlation depicting differences in the enrichment level of the SEV proteins among the clinical groups. E Gene ontology analysis (top-5 Terms) of the total SEV proteome (2306 proteins); orange, green and blue represent biological process, cellular component, and molecular function, respectively
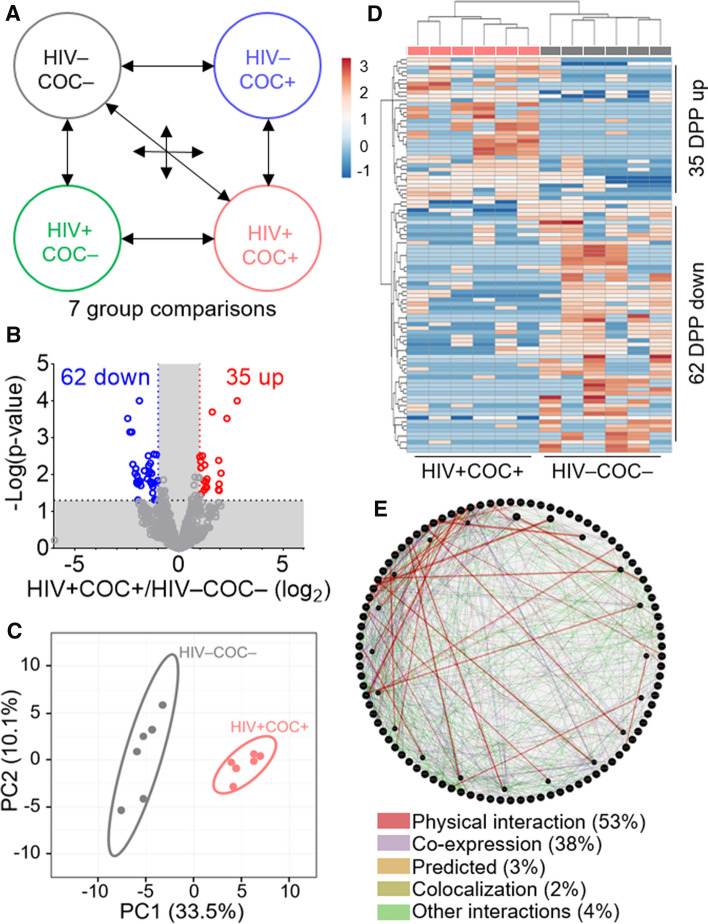
Altered SEV proteome of HIV + participants who use or do not use cocaine
To elucidate the impact of independent or combined effects of HIV infection and cocaine use on SEV proteome, we identified key markers for each clinical subgroup. To this end, we designed a 7-group comparison algorithm to facilitate the identification of modifications depicted as “Cocaine”, “HIV”, and “Comorbid” effects (Fig. 3A). The cocaine effect encompassed differentially present proteins (DPPs) in any of the following three comparisons: HIV–COC– vs HIV–COC + , HIV + COC– vs HIV + COC + and HIV + COC– vs HIV–COC + (regardless of HIV status). The HIV effect included DPPs from three comparisons: HIV–COC– vs HIV + COC–, HIV–COC + + vs HIV + COC + , and HIV- vs HIV + (regardless of cocaine use status). The comorbid effect was depicted by the DPPs resulting from HIV–COC– vs HIV + COC + group comparison. Protein enrichment signatures for the three clinical subgroups are shown in the Volcano plots (Figs. S2A, S3A, 3B). Principal Component Analysis (PCA) was employed to further unveil variation in SEV proteomes amongst the different groups. The score plots of the PCA calculated for Cocaine (Fig. S2B) and HIV (Fig. S3B) effects allowed respective separation of sample groups. However, the separation was most evident in the case of Comorbid effect (Fig. 3C). Intergroup relationship was assessed by hierarchical clustering heatmaps. There was acceptable sample clustering based on cocaine use (cocaine effect) (Fig. S2C) or HIV infection (HIV effect) (Fig. S3C), whereas a complete hierarchical sample clustering between HIV–COC– and HIV + COC + clinical subgroups was achieved for the Comorbid effect (Fig. 3D). These data suggest significant alteration of the SEV proteome by HIV infection and cocaine use, either alone or combined.
Fig. 3.
Altered SEV proteome is associated with HIV infection comorbid with cocaine use. A Group comparison scheme used to define the COC–, HIV–, and comorbid effects. B Volcano plot showing up- (red) and down- (blue) regulated proteins in the comorbid group as compared to the healthy group. C PCA plot of the 97 comorbid DPPs. Unit variance scaling is applied to rows; SVD with imputation is used to calculate principal components. X and Y axis show principal component 1 and principal component 2 that explain 33.5% and 10.1% of the total variance, respectively. Prediction ellipses are such that with probability 0.95, a new observation from the same group will fall inside the ellipse. N = 12 data points. D Hierarchical clustering heatmap of the 97 comorbid DPPs. Rows are centered; unit variance scaling is applied to rows. Both rows and columns are clustered using correlation distance and average linkage. 97 rows, 12 columns. E Protein–protein interaction (PPI) network analysis of the 97 DPPs using Genemania
For insight into the potential function of the comorbid signature proteins, we performed a protein–protein interaction (PPI) analysis of the identified DDPs in each group comparison using GeneMANIA [61]. A three-way Venn analysis showed 238, 218, and 97 DPPs for the cocaine, HIV, and comorbid effects, respectively (Fig. S2D, S3D). PPI analysis demonstrates connectivity between the DPPs in the cocaine effect (33.38% physical interactions) and the HIV effect (33.69% physical interactions) (Fig. S2E, S3E). In the Comorbid group, the connectivity between the DPPs was remarkable, with the physical interaction network reaching 52.77% of all types of interactions (Fig. 3E).
Effect of HIV infection and cocaine use on SEV miRNAome
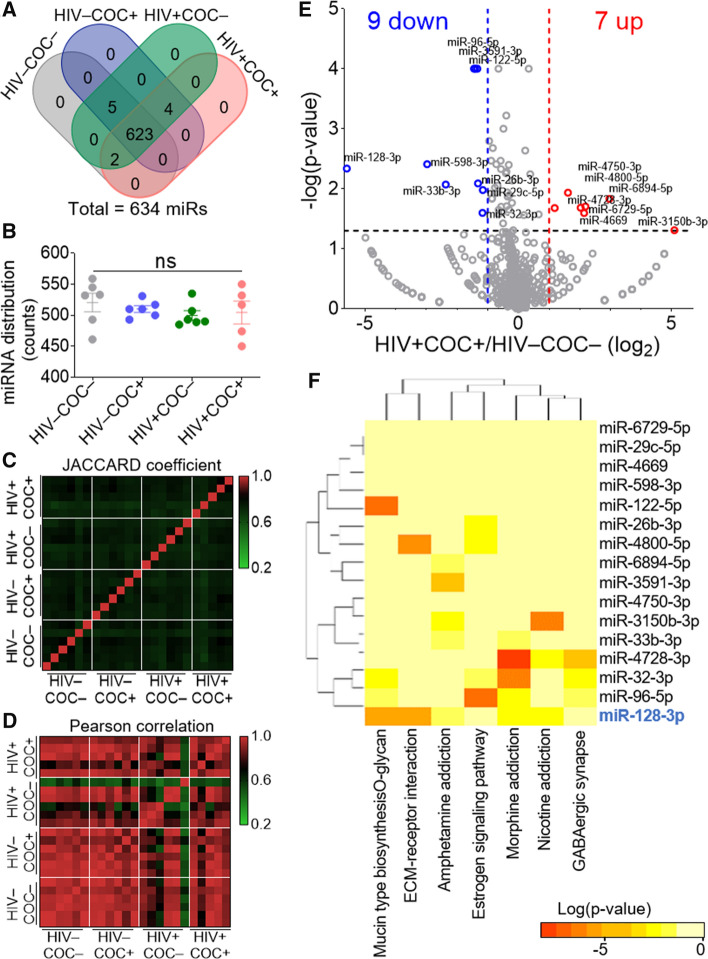
SEV miRNAome analysis was conducted using state-of-the-art low-bias sRNA-Seq technology—RealSeq® [62]. Total SEV RNA extracted from SEV isolated from 100 µL seminal plasma per sample was used. The RNA yields ranges between 241 and 1059 ng (Fig. S4A), with an A260/A280 ratio of 1.42 to 1.92 (Fig. S4B). Qubit technology (Fig. S4C) and Tape-Station RNA profiling confirm that SEV are enriched in small RNAs, although some samples contained detectable levels of larger (18S and 28S) RNAs (Fig.s S4D, E). All samples had passing-filter reads ranging from 5.5 and 10.5 million following sequencing, except one from the Comorbid group that was excluded from the subsequent analysis (Table S4). Mapping the reads to YM500v3, a human small RNA database [63], yielded 634 miRNAs (miRs) with a cutoff of 12 counts (Table S5).
Comparing miRNA distribution among samples with four-way Venn diagrams reveals more similarities than differences in miRNA signatures (Figs. 4A, B). JACCARD coefficient (Fig. 4C) and Pearson correlation (Fig. 4D) analyses of SEV miRNAome show a higher degree of similarity in miRNA levels (Pearson) compared to their distribution (JACCARD). That said, miRNAome profiles of the four groups were more similar than they were different compared to the paired proteome (Figs. 2C, D). These analyses suggest that HIV infection and/or the use of cocaine may differentially alter the enrichment patterns of proteins and miRNA in SEV.
Fig. 4.
Global analysis of SEV miRNAome and the effect of HIV infection comorbid with cocaine use. A Venn diagram showing common and unique miRNA among the clinical groups. B miRNA counts and distribution among the clinical groups. Error bars represent S.E.M. of 6 pools (n = 2) per group. Group comparison was achieved with an ordinary one-way Anova test (Tukey correction); ns, not significant. C Heatmap of the Jaccard coefficient depicting similarities and differences among clinical groups. D Pearson correlation showing differences in the enrichment level of the SEV miRNAome among the clinical groups. E Volcano plot showing up- (red) and down- (blue) regulated miRs in the Comorbid group as compared to the healthy group. F Heatmap of the union of KEGG pathways of the 16 Comorbid DP-miRs using mirPath v.3 with the default parameters
HIV infection and cocaine use endow SEV unique miRNAome enrichment pattern and regulatory network
To evaluate miRNA enrichment pattern in SEV from different clinical groups, the same strategy employed in the proteomics analysis (Fig. 3) was used. Volcano plot analysis identified a total of 15, 36, and 16 differentially present miRNAs (DP-miRs) in the “Cocaine, COC”, “HIV”, and “Comorbid” groups, respectively (Figs. 4E, S5A, S6A, B). DIANA pathway analysis [64] identified mucine type O-glycan biosynthesis, ECM–receptor interaction, Estrogen signaling pathway, as well as morphine, amphetamine, and nicotine addiction, among the top-pathways (Fig. 4F). Although several other miRs showed statistically significant differences in relative enrichment in SEV, we focused on miR-128, because it was the most downregulated miRNA in the comorbid group (Figs. 4E) that showed involvement in 5 (Mucin type O-glycan biosynthesis, ECM–receptor interaction, Amphetamine addiction, Morphine addiction, Nicotine addiction) of the seven pathways (Fig. 4F). Moreover, miR-128 plays a crucial role in normal development and various pathologies [65, 66], including mediating TNFα-induced inflammation [67], driving cocaine-induced behavioral sensitization [68], regulating bacterial [69] and viral [70–72] pathogenesis, dysregulating cytoskeletal dynamics and cell migration [73–82].
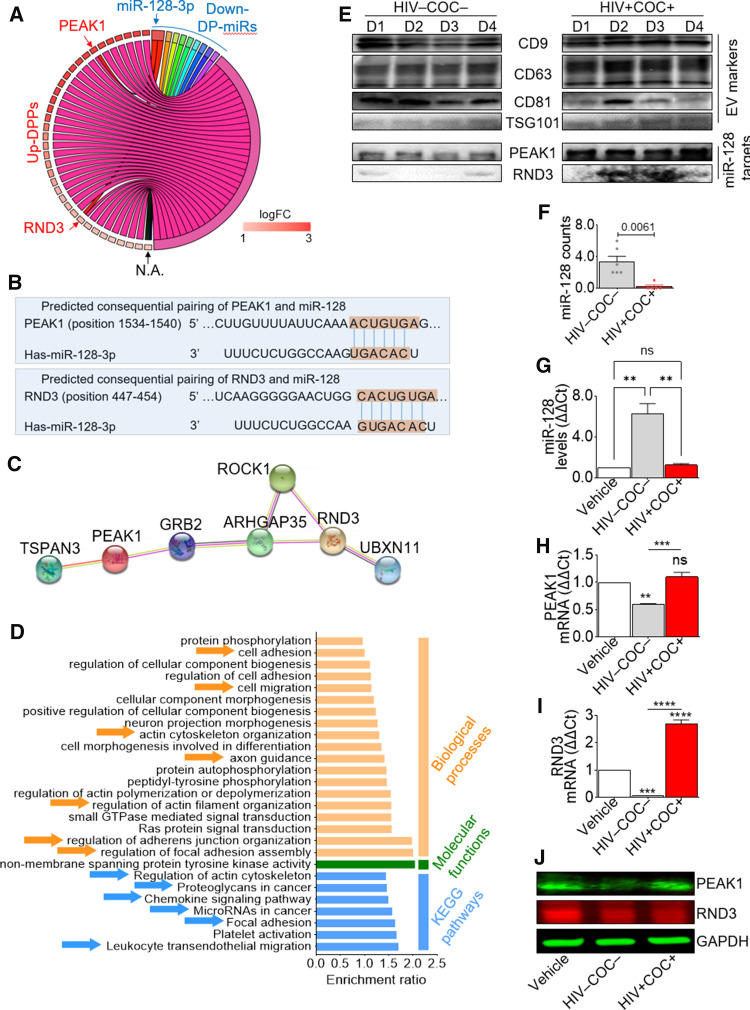
Altered miR128 network in SEV of HIV infected men comorbid with cocaine use
As identification of miR targets provides insights into their potential biological function, we determined the target genes (TGs) of the DP-miRs in the three group comparisons using miRWalk [83]. We defined a TG to be likely true if it appears in two of three databases: TargetScan, miRDB, and miRTarBase. 140, 836, and 304 TGs were identified in the COC, HIV, and Comorbid groups, respectively (Table S6). Chord diagram analysis of down-regulated miRs TGs and up-regulated DPPs in the Comorbid group identified Pseudopodium Enriched Atypical Kinase 1 (PEAK1) and Rho-related GTP-binding protein RhoE (RND3) as differential functional candidates (Fig. 5A). While PEAK1 has not been experimentally validated as miR-128 target gene, RND3 is directly targeted by miR-128 [84]. However, the 3’ UTR of the human PEAK1 and RND3 mRNA contain predicted miR-128 binding site (TargetScan 7.2) [85, 86], as shown in Fig. 5B. Comparing the human PEAK1 and RND3 3’ UTR for interspecies homology, we found that the miR-128 target sites are highly conserved among diverse species (Fig. S7). Protein–protein interaction network analysis (STRING) demonstrates an indirect connection between PEAK1 and RND3 through Rho GTPase Activating Protein 35 (ARHGAP35) and Growth Factor Receptor Bound Protein 2 (GRB2) (Fig. 5C). KEGG pathway enrichment analysis identified cell signaling pathways including platelet activation, focal adhesion, microRNAs in cancer, chemokine signaling pathway, proteoglycans in cancer, regulation of actin cytoskeleton, and leukocyte transendothelial migration (Figs. 5D). Western blot confirmed enrichment of PEAK1 and RND3 proteins in HIV + COC + SEV compared to HIV–COC– SEV, despite intra-group donor variability and similar levels of EV markers (Figs. 5E). Because of the very limited amount of SEV from all clinical groups, we were unable to address whether miR-128 is significantly varied in SEV by RT-qPCR, since a good amount of SEV is needed to isolate RNA. Nevertheless, the differential miR-128 levels as determined by the sRNA-Seq are quite pronounced (Fig. 5F).
Fig. 5.
Integrative proteome and miRNAome analysis reveals dysregulated miR128/PEAK1/RND3 network in comorbid SEV as compared to healthy. A Chord diagram showing intersection of target genes of downregulated miRs with upregulated DPPs in the Comorbid group compared to healthy. B MiRNA–mRNA duplex showing miR-128-3p binding site (seed nucleotide region) on human PEAK1 (Top) and RND3 (Bottom). C String PPI analysis showing indirect interaction of PEAK1 and RND3 through GRB2 and ARHGAP35. D Gene ontology analysis of the 7 genes identified in (C); orange, green and blue represent biological processes, molecular function, and KEGG pathways, respectively. E Western blot validation of miR-128 targets—PEAK1 and RND3 protein associated with SEV, along with EV markers. F miR-128 counts in comorbid SEV as compared to healthy. G–J Effect of SEV from the comorbid and healthy groups on endogenous monocyte G miR-128 H miR-128 target PEAK1 gene, I miR-128 target RND3 gene, as well as J PEAK1 and RND3 proteins levels. Error bars represent S.E.M. of 3 technical replicates. Experiments for panels G to I were repeated three times with similar results. Group comparison was achieved with an ordinary one-way Anova test (Tukey correction); ns not significant
To evaluate whether SEV regulate the levels of endogenous miR-128 expression and that of miR-128 predicted targets in monocytes, we analyzed the levels of miR-128, PEAK1, and RND3 after 24-h treatment of U937 cells with SEV. HIV–COC– but not HIV + COC + SEV increased the level of monocyte miR-128 (Fig. 5G) with a corresponding decrease in PEAK1 mRNA (Fig. 5H) and protein (Fig. 5J) but an increase in RND3 mRNA (Fig. 5I) and decrease in RND3 protein (Fig. 5J). Given these observations, it is likely that SEV may regulate miR-128, PEAK1, and RND3 differently and that HIV infection in the presence of cocaine may modify miRNA targets, their pathways, and functions.
Magnitude and direction of monocyte haptotaxis is dependent on the clinical source of SEV
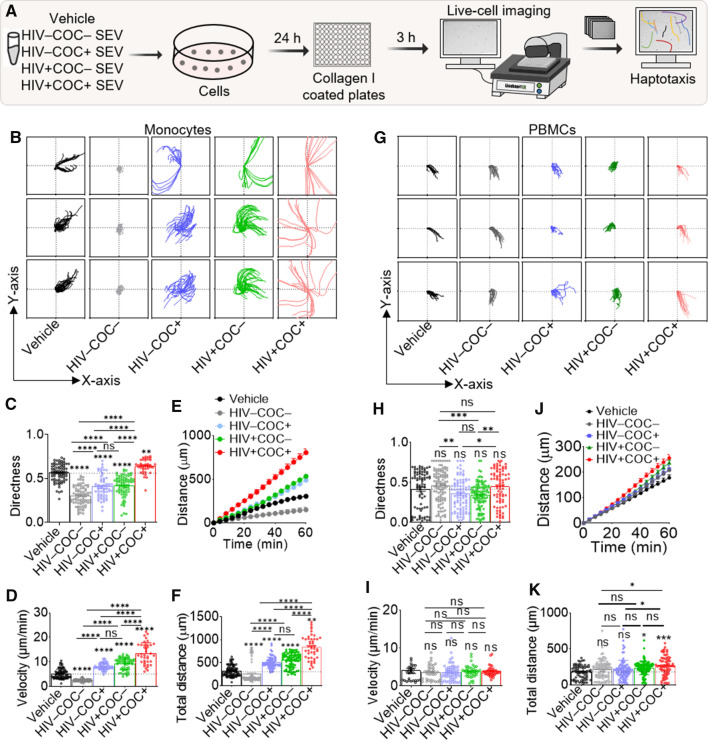
Data shown in Figs. 5E, F indicate that SEV have different enrichment patterns of the migration regulating miR-128 and its target genes, while Fig. 5D suggests KEGG pathway terms related to migration. Hence, we explored how HIV and cocaine use may affect SEV-mediated migration. To this end, we tracked single cells to study their migratory behavior in the presence of contact guidance provided by ECM networks, a process known as haptotaxis. U937 cells cultured under steady state (vehicle, PBS) or in the presence of HIV–COC–, HIV + COC + SEV for 24 h were seeded atop collagen-coated wells (Fig. 6A). Individual monocyte tracks for every 4 min period were calculated and sequentially plotted (Figs. 6B). Tracks were analyzed to extract values for directedness (Fig. 6C), velocity (Fig. 6D), migration kinetics (Fig. 6E) and cumulative distance (Fig. 6F). The data revealed that compared to steady-state haptotaxis, monocytes exhibit a rapid and more directional haptotactic migration pattern in the presence of HIV + COC + SEV that is distinct from decreased haptotaxis in the presence of HIV–COC– SEV (Figs. 6C–F).
Fig. 6.
SEV from HIV + COC + increase the magnitude and direction of leukocyte haptotaxis. A Schematic of workflow for SEV haptotaxis assay. Briefly, U937s (0.5 M cells) were treated with either Vehicle, or different donor group SEVs (100 µg) for 24 h. Equivalent numbers of viable cells were plated on collagen type I coated plates for haptotaxis assay. B Representative plots of single monocyte tracks following treatment with Vehicle or SEV from the different clinical groups. C–F Plots for monocyte directedness, average velocity, kinetic distance migrated, and total distance migrated, respectively. G Representative plots of single PBMC tracks treated with Vehicle or SEV from the different clinical groups. H–K Plots for PBMC directedness, average velocity, kinetic distance migrated, and total distance migrated, respectively. The plots C–F and H–K were calculated for 1 h at 9 equidistant fields of view per well, in triplicates. Experiment was repeated 3 times for monocytes and 2 times for PBMCs, each from two independent donors. Ordinary one-way ANOVA test (Dunnett’s correction) was used to determine the differences between the treated groups as compared to vehicle treatment. Two-tailed Welch’s t test was used to compare the differences between SEV treatments. ****p < .001, ***p < .005, **p < .01, * p < .05, ns not significant
To confirm the reported observation, peripheral blood mononuclear cells (PBMCs) were treated with the four SEV groups under the same conditions as for monocytes (Fig. 6A). Individual tracks (Figs. 6G) show that SEV had varied effects on PBMC directedness (Fig. 6H) and haptotaxis speed (Figs. 6I). However, there was a consistent increase in the migration and cumulative distance that PBMCs traveled when in the presence of HIV + COC + SEV (Figs. 6J, K) compared to HIV–COC– or vehicle.
The fact that SEV-mediated regulation of haptotaxis also occurred with HIV–COC + and HIV + COC– SEV in monocytes (Figs. 6B–F) and PBMCs (Figs. 6G–K), although to a lesser extent, suggest that both HIV and cocaine are contributory to the combined comorbid effect. The functional effects of the different SEV on haptotaxis are not due to cell death, since cell viability were similar across treatments (Fig. S8). These data indicate that donor HIV and cocaine use status alter the ability of SEV to regulate haptotaxis speed and directionality in monocytes and PBMCs.
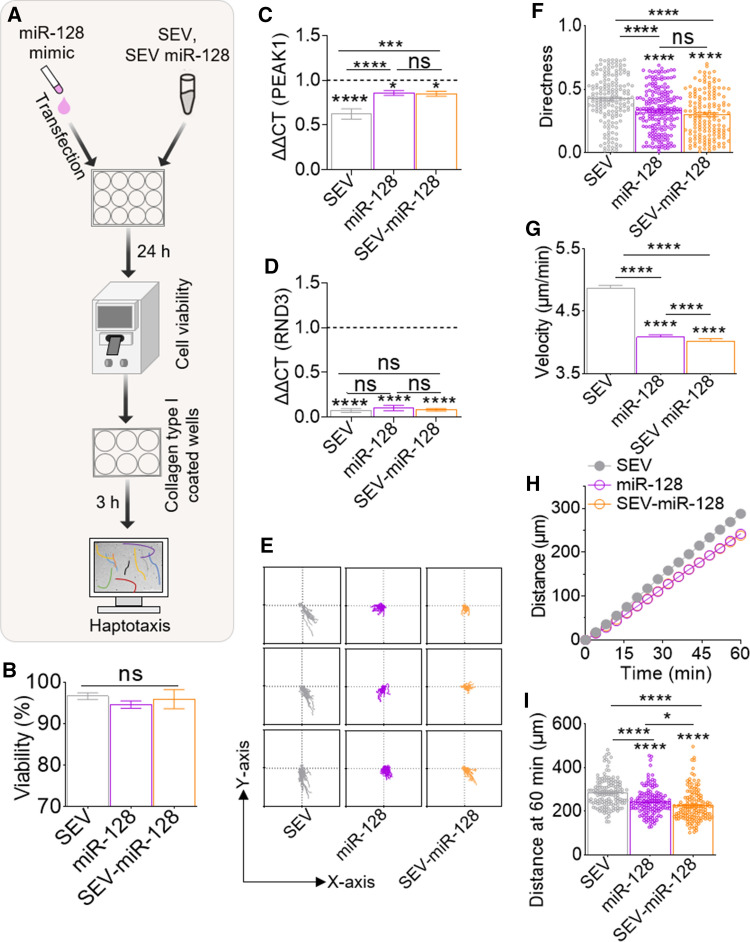
SEV-loaded with miR-128 mimic potentiate decreased monocyte haptotaxis
Data presented thus far showed differential (i) enrichment of miR-128 and its TGs in HIV–COC– and HIV + COC + SEV (Fig. 5E, F), (ii) alteration of endogenous levels of miR-128 and its TGs in monocytes by SEV (Fig. 5G–J), as well as (iii) regulation of distinct haptotactic migration by HIV–COC– and HIV + COC + SEV (Fig. 6B–K). Based on these data, we specifically investigated the relationship between SEV-associated miR-128 and haptotaxis. To this end, we loaded healthy donor SEV with miR-128 mimic (SEV-miR-128). Briefly, 100 µg of SEV from healthy donors was transfected with 150 pmol of miR-128-3p and purified via filtration through a 10-kDa filter. The loaded SEV was used to treat monocytes. Cells were also treated with various controls, including SEV alone, miR128 alone (Fig. 7A), or vehicle PBS and All-Star Negative Control, an siRNA with no homology to any known gene (miR-CTL), Figure S9). None of the treatments had any noticeable effect on monocyte viability (Fig. 7B, S9B). However, SEV-miR-128 decreased the levels of PEAK1 and RND3 as expected (Fig. 7C, D). Following single cell tracking, haptotactic measures including cell directedness, velocity, migration kinetics, and cumulative traveled distance were calculated. As expected, miR-CTL did not affect haptotactic measures compared to vehicle PBS, except for directness (Fig. S9E–I). However, miR-128 alone or encased within SEV halted monocyte migration (Fig. 7E) as indicated by a decreased directedness (Fig. 7F), velocity (Fig. 7G) and total migrated distance (Fig. 7H–I), compared to SEV alone. The trajectories of the cells treated with SEV-miR-128 show an almost complete halt, which appears similar to miR-128 alone compared to SEV alone (Fig. 7H). These findings, which suggest that perturbed haptotactic velocity and distance might be regulated by exmiR-128, further validate the results of SEV regulated haptotaxis in Fig. 6, where monocytes and PBMCs migration velocity and distance were increased by miR-128 depleted HIV + COC + SEV but decreased by miR-128 enriched HIV–COC– SEV.
Fig. 7.
SEV-loaded miR-128 mimic potentiates decreased monocyte haptotaxis. A Schematic of workflow for SEV-miR-128 mimic studies. Briefly, monocytes were either transfected with miR-128 mimic or treated with HIV–COC– SEVs loaded with miR-128 mimic. Following 24 h incubation, cell viability was assessed, and cells were plated onto collagen I coated plates for haptotaxis assay. B Viability of monocytes treated with miR-128, SEV, or SEV-miR-128. C–D Gene expression for miR-128 target genes. E Representative plots of single monocyte tracks following treatment with miR-128, SEV, or SEVmiR-128. F–I Plots for monocyte directness, average velocity, kinetic distance migrated, and total distance migrated, respectively. The plots were calculated for 1 h at 9 equidistant fields of view per well, in triplicates. Ordinary one-way ANOVA test (Dunnett’s correction) was used to determine the differences between the treated groups as compared to vehicle treatment. Two-tailed Welch’s t test was used to compare the differences between SEV treatments. ****p < .001, ***p < .005, **p < .01, *p < .05, ns not significant
Discussion
In this study, we employed an integrative multi-omics systems biology (proteomics and sRNA-Seq), along with ECM-guided single cell migration assay, to explore the role of extracellular vesicles in monocyte haptotaxis. To this end, we used omics studies to characterize the cargo composition of SEV from diverse clinical groups and then set up in vitro haptotaxis experiments to investigate how HIV/cocaine-induced changes in SEV proteome and miRNAome affect haptotaxis. In the proteomics analysis, we included proteins with one unique peptide, allowing identification of numerous low-expressing cell surface proteins, such as CD151, CD180, CD200 and CD99L2. This strategy is not uncommon in proteomics studies [87, 88] and it was estimated that 30–50% of the proteome are identified with one unique peptide [89]. Of note, CD99L2 is an adhesion molecule that plays a key role during inflammation-mediated leukocyte extravasation [90, 91]. Our integrative omics studies revealed that miR-128 was relatively high in HIV–COC– SEV, low in HIV + COC– (Anova, p value = 0.0246), and lowest in HIV + COC + (Anova, p value = 0.0065), suggesting that HIV infection and cocaine use resulted in the release of SEV with decreased miR-128. By analyzing hundreds of monocytes and PBMC haptotactic trajectories, we determined that the directionality of the cells, their corresponding velocity, and migration distance are increased in the presence of HIV + COC + SEV but decreased when HIV–COC– SEV are present, suggesting that cell migration is rapid and mostly directional in the presence of HIV + COC + SEV.
Measurements of monocytes and PBMC trajectories, velocities, and distance revealed substantial differences in haptotactic response to SEV. While monocytes haptotactic directedness is significantly different in the presence of the different SEV (Fig. 6F), PBMC directionality was donor-dependent (Fig. 6K). However, the cumulative distance migrated increased in both monocytes (Fig. 6I) and PBMCs (Fig. 6N) in the presence of HIV + COC + SEV, irrespective of donor variability (PBMCs). The exact mechanism underlying haptotactic migration differences between these cells in response to SEV is unknown. Whereas monocytes are homogenous, PBMCs consist of a mixture of different cell types, including B, T, NK, dendritic cells, and monocytes, the percentages of these cell types within PBMCs may differ between donors. All of these differences may have different haptotactic responses to the SEV and SEV-associated miR-128. It is possible that, in addition to the above cell type-specific differences, cells may differently interact with possible topographic features to influence the organization of the actin cytoskeleton and focal adhesions to alter haptotactic responses.
Functionalizing HIV–COC– SEV with miR-128 mimic potentiates decrease in the velocity and cumulative migration distance of cells (Fig. 7). MiR-128 is an intronic miRNA encoded by two distinct genes, miR-128–1 and miR-128–2. The host gene of miR-128 is the regulator of calmodulin signaling (Rcs), also known as Arpp21, and Rac is essential for mediating dopamine transmission [92]. The brain, frontal cortex, differentiating DA neurons [93], and the male reproductive tract (MRT) [94, 95] all express miR-128. Given its tissue-specific expression, miR-128 plays a broad and crucial role in various pathologies. In the central nervous system (CNS), miR-128 plays a role in development [65]. Furthermore, miR-128 expression is elevated in Alzheimer’s disease brain [66], where it regulates motor activity and neuronal excitability through the suppression of various ion channels and ERK1/2 signaling network [66]. In viral pathogenesis, miR-128 is an interferon-inducible miRNA that enhances HIV-1 Replication by repressing nuclear import factor (TNPO1) mRNA expression [96]. In addition, miR-128 is a Tat-responsive miR upregulated in Tat-treated primary rat cortical neurons to control neuronal activity [70]. Furthermore, in the MRT of stallions, miR-128 is decreased by equine arteritis virus [71]. Downregulated miR-128 was associated with increased CXCL16 and long term persistence of equine arteritis virus [71], while the synthesis of human rhinovirus (HRV-1B) RNA was increased upon suppression of miR-128 [72], an indication of antiviral function. Furthermore, Salmonella enterica serovar enteritidis proteins upregulate miR-128 expression, resulting in decreased M-CSF secretion and impaired macrophage recruitment [69]. Interestingly, miR-128 responds to cocaine use, since it contributes to methamphetamine-induced behavioral sensitization by controlling neuroplasticity [68].
The identification of miR-128 in SEV and its reduced enrichment in HIV + COC + SEV is remarkable and may suggest potential brain-gonadal crosstalk. Indeed, gonadal dysfunction may be linked to physiological, developmental, pathological (viral infections) problems of the gonad and intrinsic processes in the CNS, including chemosignals that affect reproduction [97]. Hormonal and neurochemical regulation involved in reproduction may be regulated by brain-enriched miRNAs. It has been shown that testosterone treatment increased the expression of miR-128, which in turn decreased catalytic function of CYP2D6 in a cell type-specific manner [98]. On the other hand, orchiectomy (surgical removal of the testicles), which decreases testosterone levels, also decreased the levels of miR-128 in the hippocampus of males [98]. These studies indicate that hormones, such as androgens regulate miRNAs and may regulate the levels of miR-128.
The findings of this study are consistent with the observations that EVs constitute an important but complex factor in disease pathogenesis and co-morbidities of cocaine use. Indeed, studies have shown that miRNAs are encapsulated in EVs, and cocaine use regulate EV-associated miRNAome [99–101]. Upon cellular internalization of EV-containing miRNAs, the miRNAs act as endogenous miRNA to alter the levels of target gene expression [99]. In our study, we observed that HIV–COC– SEV are enriched in miR-128 (Figs. 4E) with an attendant decrease in protein levels of the miR-128 target genes PEAK1 and RND3 (Figs. 5E). This inverse relationship between miR-128 and PEAK1/RND3 was validated in cells treated with HIV–COC– SEV (Figs. 5G–J). The multifaceted function of SEV is further confirmed by the observation that SEV from different clinical backgrounds differentially regulate miR-128 and its targets. The finding that HIV–COC– but not HIV + COC + SEV increased miR-128, reduced PEAK1 and RND3 mRNA and protein is interesting. However, this is not surprising, because SEV contain other cargos and miR-128 have been shown to modulate protein levels without affecting mRNA target [102]. Hence, it is possible that the observed effect may be mediated by other SEV cargoes. Noteworthy, exogenous miR-128 mimic decreased PEAK1 and RND3 mRNA (Figs. 7C, D), further indicating that PEAK1 and RND3 are a part of miR-128 network involved in regulating monocyte haptotaxis.
Our results provide a molecular view of SEV-mediated higher order interactions in cellular behavior and how HIV infection and cocaine use may regulate SEV cargo composition and function. What we did not assess in this study is the effector of miR-128 function, the extent to which such effectors influenced monocyte haptotaxis in the context of HIV infection/cocaine use, and downstream signaling cascades involved. Though beyond the scope of the current studies, these experiments are necessary for complete understanding of miR-128 and its role in EV-mediated cellular behavior. While the inclusion criterion for cocaine use was the use of cocaine, it is noteworthy that study participants reported using other drugs. Hence, it is plausible that “COC effect” observation may be mediated by multiple illicit drugs. Studies focused on cocaine alone are needed to clarify the contribution of cocaine. Animal models of HIV/AIDs, including the non-human primates, are ideal for such studies.
The distinct haptotactic responses by cells treated with different SEV (HIV + COC + and HIV–COC–) is indicative that EVs and their exmiR networks may provide cells divergent haptotactic guidance in the absence of chemotactic cues, both under physiological and pathophysiological conditions. HIV + participants used in this study were ART-naïve. Thus, the findings are more relevant to new infections prior to initiation of ART or to people living with HIV (PLWH) who are unable to access ART or to stay on therapy. In these groups of people, monocytes and macrophages are important drivers of pathogenesis and progression to AIDS. The finding that SEV (via the cargos) from different clinical groups are capable of reprograming the function of monocytes should awaken the reappraisal of monocytes and their circulation dynamics in and out of different tissues. For example, altered levels of miR-128 in HIV + COC + SEV may have implications for the pathogenesis of viruses inhibited by SEV, including HIV [4, 5] and ZIKV [103]. A reduction in miR-128 may promote trafficking of virus infected monocytes from the initial site of infection to various anatomic sites, such as the brain, lung, and lymph nodes. In addition, modification of SEV cargo in general and SEV-associated miR-128 may epigenetic modifications in paternally inherited characteristics in offspring [104] in both placental and brain tissues during development [105].
With this study, a new framework exists to contextualize and evaluate the significance of monocytes in pathogenesis of infectious agents comorbid with the epidemic of drug use. It is plausible that EV-mediated distinct migratory behaviors in response to diverse guidance cues may be physiologically or pathophysiologically relevant in shaping fertilization, development, host response to infection, or in the recruitment of immune cells to the site of inflammation.
Materials and methods
Ethics and semen samples
This study was conducted according to University regulations approved by Stony Brook University Institutional Review Boards (IRB # 201,608,703) using de-identified human specimens obtained through the Multicenter AIDS Cohort Study (MACS) [52]. 64 specimens obtained from 4 clinical groups (16 per group), HIV–COC–, HIV–COC + , HIV + COC–, and HIV + COC + were used. The selection criteria and the clinical characteristics were recently described [40] and further discussed in the Results section under study design. The semen samples were received on dry ice and stored at − 80 °C until analysis.
Cells and reagents
Human U937 monocyte-like cells were obtained from American Type Culture Collection (ATCC) and maintained in complete RPMI media. The complete media was supplemented with 10% EV-depleted FBS, 1% penicillin–streptomycin, 1 µg/mL amphotericin B, 2 mM sodium pyruvate, 1% glutamate, and 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer at pH 8.
EV isolation
The EVs were isolated from the semen specimens by differential centrifugation and PPLC-based size exclusion chromatography (SEC) as previously described [40]. Briefly, samples were liquefied at room temperature for 30 min, centrifuged at 2000×g for 10 min and 10000×g for 30 min to remove cellular debris and large vesicles. Clarified samples were pooled into six separate pools (n = 2 samples, 100 µL/sample) for each group. EVs were purified using a gravity-packed sephadex G-50 resin into 22 cm × 1 cm Econo-column. Elution was achieved by gravity using Phosphate Buffered Saline (PBS, Corning, NY, USA). Fractions of 200 µL were collected, and elution profiles were determined by absorbance measurements at 280 nm and 600 nm. The void peak which contained the SEVs was collected, pooled, and stored in aliquots at – 80 °C. The protein content was measured by the Bradford Assay (Bio-Rad, Hercules, CA, USA).
Nanoparticle tracking analysis (NTA)
SEV size, concentration, and zeta potential were measured using ZetaView PMX 110 (Particle Metrix, Mebane, NC, USA) and the corresponding software ZetaView v8.04.02, as previously described [54].
Silver stain and Western blotting
10 µg of SEV proteins or 30 µg of SEV-treated cell lysates were loaded on pre-cast 4–20% polyacrylamide Criterion gels (BioRad). Gels were either stained using Silver Stain Plus Kit (BioRad) following the manufacturer’s protocol or blotted onto a PVDF membrane. The blots were blocked with 5% BSA in 1X TBST buffer and incubated with the primary antibody at 4 °C overnight. The blot was rinsed with 1X TBST 3–5 times and incubated with the appropriate secondary antibody for 1 h at room temperature, then imaged (Odyssey infrared imaging system, LI-COR). Protein band intensities were quantified (ImageJ, NIH, Bethsada, MD).
Protein digestion for proteomics
Prior to LC–MS/MS analysis, protein samples were denatured in 8 M urea and 50 mM Tris–HCl, pH 8.0, reduced with 10 mM TCEP for 60 min at RT, alkylated with 2 mM iodoacetamide for 60 min at RT, and then diluted to 2 M urea with 50 mM Tris–HCl, pH 8.0. Trypsin Gold (Promega) was added to the mixture (1:25 w/w enzyme to protein ratio) before overnight digestion (18 h, 37 °C). The tryptic peptides were desalted using Pierce C18 spin columns (Thermo Fischer Scientific) at RT. Peptides were eluted with 80% acetonitrile and 0.1% formic acid (FA) and then dried completely on a SpeedVac Concentrator.
LC–MS/MS analysis
40 µg of trypsinized peptides were resuspended in 5 μL of 0.5% FA and loaded onto a three-phase MudPIT column (150 μm × 2 cm C18 resin, 150 μm × 4 cm SCX resin, filter union, and 100 μm × 12 cm C18 resin) as described previously [1]. A ten-step MudPIT protocol was used for the LC–MS/MS analysis on an Eksigent™ AS-1 autosampler and Eksigent™ nano-LC Ultra 2D pump inline with a Thermo Fischer Orbitrap LTQ XL linear ion trap mass spectrometer with a nanospray source. Each step consisted of 0 mm, 25 mM, 50 mM, 100 mM, 150 mM, 200 mM, 300 mM, 500 mM, 750 mM, and 1000 mM ammonium acetate with each salt pulse followed by a 120-min acetonitrile gradient 5–50% B [Buffer A: 0.1% FA; Buffer B: 0.1% FA in acetonitrile]) MS data acquisition was done in a data-dependent “Top-5” method (a survey FTMS scan [res. 30,000] followed by five data-dependent IT scans for the five consequent most abundant ions). The general mass spectrometric settings were as follows: spray voltage, 2.4 kV; no sheath and no auxiliary gas flow; ion transfer tube temperature, 200 °C; CID fragmentation (for MS/MS), 35% normalized collision energy; activation q = 0.25; activation time, 30 ms. The minimal threshold for the dependent scans was set to 1000 counts, and a dynamic exclusion list was used with the following settings: repeat count of 1, repeat duration of 30 s, exclusion list size of 500, exclusion duration of 90 s. We adjusted the area under the curve (AUC) values for each protein (as calculated by the PEAKS software) to the corresponding spectral count (SpC) by nullifying AUC values for proteins with no detectable SpC [1], and then log10-normalized these values. An in-house sequence database was constructed using the Swiss-Prot UniProt Human non-redundant, manually annotated database (up000005640, version release 2018_04) as the reference database (https://www.uniprot.org/uniprot), to which 381 HIV protein sequences (from all clinical isolates available in Uniprot) were concatenated.
Total RNA isolation
Total RNA from SEV was isolated from 1 mL of SEV (corresponding to ~ 100 µL seminal plasma) using miRNeasy plasma kit (Qiagen), with the optional on-column DNase-I digestion step. Total RNA was eluted in 25 µL RNase-free water once and once with the eluate. RNA quality control was assessed by Nanodrop and Agilent Bioanalyzer prior to sequencing.
Library preparation and sRNA sequencing
Libraries were amplified by 20 cycles of PCR. Libraries were sequenced in one NextSeq 550 run with the NextSeq 500/550 High Output Kit v2.5 (75 cycles), sequencing was done with Single End 75 nt reads and dual 6 nt indexes. Libraries were loaded at 1.5 pM and sequenced with a RealSeq Biosciences (Santa Cruz, CA) custom sequencing primer for read one; 5% PhiX control was used.
Bioinformatics and RNA identification pipeline
For adapter trimming, raw sequencing files for each sample were merged using the ‘cat’ via the command line to generate a single fastq file per sample. The adapter sequence was trimmed, and reads with inserts smaller than 15 nt were removed. Trimmed reads were aligned to two references hg19 and YM500v3, the miRBase data set for human miRs using bowtie. Samtools 1.7, using htslib 1.7–2, was used to convert and sort the SAM files into sorted BAM files. T ‘BuildBamIndex’ picard-tool via the PicardCommandLine was used to generate an index and convert BAMs to reads files. These files contained the name of the object aligned to, and the number of raw reads that were aligned. A count matrix was made using a proprietary Python program.
For miRs differential presence analysis, we used as a cutoff the sum of reads, which was defined as equal or larger than the number of samples being compared per group times two. Thus, for a miR to be included in the analysis the sum of reads should be ≥ 12. The row sum filtration is important, because it accounts for low expressing miRs that may be consistently counted in one sample but not, or just a few times, in others. In this case, it is important to keep zero value counts, because they may have biological relevance. Subsequently, the read counts were log-10 normalized and two-way ANOVA comparisons between different groups performed in Prism software. The false discovery rate (FDR) was controlled using the method of Benjamini and Hochberg and was set to < 0.05. Under these parameters, only a few hits withstood the analysis, prompting us to set a lesser stringent cutoff as follows: -log(P value) < 0.05 and fold change (FC) of > 2.
Prediction of the DPPs–miRNA interactions
DP-miRs target genes were predicted using miRWalk 3.0 (http://mirwalk.umm.uni-heidelberg.de/) [83], integrating the prediction results from at least two of the three available databases TargetScan [106], miRDB [107], and miRTarBase [108]. The default score ≥ 0.95 was considered as the cutoff criterion for the prediction analysis in miRWalk. Predicted target genes of the upregulated DP-miRs were intersected with the downregulated DPPs from the corresponding group comparison and vice versa. The overlap of these intersections were further analyzed for protein–protein interaction using STRING (https://string-db.org/) and GENEMANIA (http://genemania.org), and for gene ontology and signaling pathways using WEBGESTALT (http://www.webgestalt.org/).
Data visualization, PPI analysis, and pathway identification
Venny, InteractivVenn, heatmapper, ClustalVis, Cytoscape software (version 3.40) GENEMANIA, STRING, and WEBGASTALT were used to calculate and visualize the regulatory networks. R Studio (version 4.0.1) was used to generate the Chord diagrams using the GOPlot package [109].
Transfection of miR-128-3p mimic
150 pmol of synthetic LNA-based Syn-hsa-miR-128-3p miScript miRNA Mimic (miR-128, Qiagen Cat No./ID: MSY0000424|S2) or All-Star-Negative Control (miR-CTL, Qiagen Cat No./ID: 1,027,281) were diluted in 100 µL Opti-MEM medium to which 3 µL of Hiperfect transfection reagent (Qiagen) diluted in equal volume of Opti-MEM were added, and the mixture was vortexed. After 10 min incubation at room temperature to allow for the formation of transfection complexes, the mixture was added dropwise onto 0.5 million of U937 cells, cultured in a 12-well plate in 1 mL of cRPMI. 24 h later, cells were assessed for viability and harvested for subsequent gene expression (mRNA and protein) analysis and single cell migration.
Loading of SEV with miR-128-3p mimic
miR-128-3p mimic (150 pmol) was diluted in 100 µL of Opti-MEM medium. 3 µL of Hiperfect transfection reagent was added to the mixture and vortexed. Samples were incubated for 10 min at room temperature to allow for the formation of transfection complexes. 100 µg of SEV from healthy donors (HIV–COC–) were diluted in 100 µL 1X PBS, added to the complexes, and mixed by inversion 3 times. The mixture was incubated at room temperature for 1 h. Following incubation, SEV were purified using 3 times filtration through a 10-kDa filter with washing by 1X PBS.
Treating cells with SEV or SEV loaded with miR-128 mimic
0.5 million cells that were at exponential growth phase were plated per well in a 12-well plate in 1 mL of EV-free cRPMI. 100 µg of SEV were added dropwise onto cells and incubated for 24 h.
Single cell tracking
Following incubation with respective treatments, cells were seeded (2,000/well) in a 96-well plate coated with 50 µg/mL type I collagen (Bovine, Corning). Cells were then allowed to adhere for 3 h at 37 °C. Kinetic images were acquired using the Lionheart FX automated microscope. 10× magnification brightfield images were captured at 4 min intervals for 1 h at 9 equidistant fields of view per well. Image processing was performed using Gen5 ImagePrime software. Kinetic frame alignment was applied to improve image quality and remove X–Y movement between frames that occurs naturally during image capture. All cells present in the field of view for the entirety of the kinetic read were tracked using the Manual Tracking plugin for ImageJ. Cumulative distance migrated () and average velocity (), where i = index of the single cell, were calculated using the plugin. Directionality (Di) was calculated by comparing the euclidian distance () and the accumulated distance (di,accum) between the starting point and the endpoint of a migrating cell ().
Gene expression
Equivalent amount of RNA from either cells or SEV were used for cDNA synthesis using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermofisher, Grand Island, NY, USA). The cDNA was then used for RT-qPCR assay. The thermal cycler program was setup in a 7500 FAST real-time PCR system (Applied Biosystems, Thermofisher, Grand Island, NY, USA). The fold change in gene expression was calculated using the standard ∆∆CT method. The primers used are presented in Table S6.
Statistical analysis
GraphPad Prism (Version 9.2.0) was used to plot all graphs and perform all statistical analyses. As highlighted in Fig. 1A, the SEV characterization, proteomics and sRNA-Seq analyses (Figs. 1C–J, 2B–D, 3B–D, 4B–E) were conducted using 6 biological pools (n = 2) per group, and an ordinary one-way Anova test (Tukey correction) was used for group comparisons. For validation experiments, samples from individual donors (n = 4 per group) were used either individually (Fig. 5E) or pooled (Fig. 5G–J, 6B–K). PBMC treatments used two healthy donors. All single cell tracking experiments were conducted using randomly selected equidistant field of view per well from 3 wells and the experiments were repeated two (PBMCs) or three times for monocytes.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Institute on Drug Abuse (NIDA) grants DA042348, DA050169 and DA053643 (to CMO). Our gratitude goes to Bryson Okeoma of Stony Brook University for critical review of this manuscript. Data in this manuscript were collected by the Multicenter AIDS Cohort Study (MACS) with centers at Baltimore (U01-AI35042): The Johns Hopkins University Bloomberg School of Public Health: Joseph B. Margolick (PI), Todd Brown (PI), Jay Bream, Adrian Dobs, Michelle Estrella, W. David Hardy, Lisette Johnson-Hill, Sean Leng, Anne Monroe, Cynthia Munro, Michael W. Plankey, Wendy Post, Ned Sacktor, Jennifer Schrack, Chloe Thio; Chicago (U01-AI35039): Feinberg School of Medicine, Northwestern University, and Cook County Bureau of Health Services: Steven M. Wolinsky (PI), Sheila Badri, Eun-Young Kim, Dana Gabuzda, Frank J. Palella, Jr., Sudhir Penugonda, John P. Phair, Susheel Reddy, Matthew Stephens, Linda Teplin; Los Angeles (U01-AI35040): University of California, UCLA Schools of Public Health and Medicine: Roger Detels (PI), Otoniel Martínez-Maza (PI), Otto Yang (Co-PI), Peter Anton, Robert Bolan, Elizabeth Breen, Anthony Butch, Shehnaz Hussain, Beth Jamieson, John Oishi, Harry Vinters, Dorothy Wiley, Mallory Witt, Stephen Young, Zuo Feng Zhang; Pittsburgh (U01-AI35041): University of Pittsburgh, Graduate School of Public Health: Charles R. Rinaldo (PI), Lawrence A. Kingsley (PI), Jeremy J. Martinson (PI), James T. Becker, Phalguni Gupta, Kenneth Ho, Susan Koletar, John W. Mellors, Anthony J. Silvestre, Ronald D. Stall; Data Coordinating Center (UM1-AI35043): The Johns Hopkins University Bloomberg School of Public Health: Lisa P. Jacobson (PI), Gypsyamber D’Souza (PI), Alison Abraham, Keri Althoff, Michael Collaco, Priya Duggal, Sabina Haberlen, Eithne Keelaghan, Heather McKay, Alvaro Muñoz, Derek Ng, Anne Rostich, Eric C. Seaberg, Sol Su, Pamela Surkan, Nicholas Wada. Institute of Allergy and Infectious Diseases: Robin E. Huebner; National Cancer Institute: Geraldina Dominguez. The MACS is funded primarily by the National Institute of Allergy and Infectious Diseases (NIAID), with additional co-funding from the National Cancer Institute (NCI), the National Institute on Drug Abuse (NIDA), and the National Institute of Mental Health (NIMH). Targeted supplemental funding for specific projects was also provided by the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute on Deafness and Communication Disorders (NIDCD). MACS data collection is also supported by UL1-TR001079 (JHU ICTR) from the National Center for Advancing Translational Sciences (NCATS) a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH), Johns Hopkins ICTR, or NCATS. The MACS website is located at http://aidscohortstudy.org/.
Author contributions
Conceptualization, CMO; methodology, CMO, HK, SK, YL, and NS; validation, HK, SK, YL, and NS; formal analysis, CMO, HK, SK, YL, and NS; investigation, CMO, HK, SK, YL, NS, VP, and SP; resources, CMO, CD, EK, JM, HM, ME, JBM, and JTS; data curation, HK; writing—original draft preparation, CMO, HK, SK, YL, and NS; writing—review and editing, CMO, HK, SK, YL, VP, SP, and NS. CD, EK, JM, HM, ME, JBM, and JTS. supervision, CMO; project administration, CMO; funding acquisition, CMO. All authors participated in manuscript preparation and approved the final version of the manuscript.
Funding
This research was funded by the National Institute on Drug Abuse (NIDA), grants DA042348, DA050169 and DA053643 to C.M.O
Availability of data and materials
The mass spectrometry proteomics data sets supporting the conclusions of this article have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD020487. The sRNA-Seq data sets are included within the article and its additional files.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This study was conducted according to University regulations approved by Stony Brook University Institutional Review Boards (IRB # 201608703) using de-identified human specimens obtained through the Multicenter AIDS Cohort Study (MACS).
Consent to participate
Individuals who met the inclusion and exhibit none of the exclusion criteria and who gave written informed consent were included in the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kaddour H, Lyu Y, Welch JL, Paromov V, Mandape SN, Sakhare SS, Pandhare J, Stapleton JT, Pratap S, Dash C, Okeoma CM. Proteomics profiling of autologous blood and semen exosomes from HIV-infected and uninfected individuals reveals compositional and functional variabilities. Mol Cell Proteomics. 2020;19:78–100. doi: 10.1074/mcp.RA119.001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaddour H, Lyu Y, Shouman N, Mohan M, Okeoma CM. Development of novel high-resolution size-guided turbidimetry-enabled particle purification liquid chromatography (PPLC): extracellular vesicles and membraneless condensates in focus. Int J Mol Sci. 2020;21:5361. doi: 10.3390/ijms21155361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M, Hladik F. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madison MN, Jones PH, Okeoma CM. Exosomes in human semen restrict HIV-1 transmission by vaginal cells and block intravaginal replication of LP-BM5 murine AIDS virus complex. Virology. 2015;482:189–201. doi: 10.1016/j.virol.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madison MN, Roller RJ, Okeoma CM. Human semen contains exosomes with potent anti-HIV-1 activity. Retrovirology. 2014;11:102. doi: 10.1186/s12977-014-0102-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welch JL, Kaddour H, Schlievert PM, Stapleton JT, Okeoma CM. Semen exosomes promote transcriptional silencing of HIV-1 by disrupting NF-kB/Sp1/Tat circuitry. J Virol. 2018;92(21):e00731. doi: 10.1128/JVI.00731-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch JL, Kaddour H, Winchester L, Fletcher CV, Stapleton JT, Okeoma CM. Semen extracellular vesicles from HIV-1-infected individuals inhibit HIV-1 replication In Vitro, and extracellular vesicles carry antiretroviral drugs In Vivo. J Acquir Immune Defic Syndr. 2020;83:90–98. doi: 10.1097/QAI.0000000000002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch JL, Kaufman TM, Stapleton JT, Okeoma CM. Semen exosomes inhibit HIV infection and HIV-induced proinflammatory cytokine production independent of the activation state of primary lymphocytes. FEBS Lett. 2020;594:695–709. doi: 10.1002/1873-3468.13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welch JL, Madison MN, Margolick JB, Galvin S, Gupta P, Martinez-Maza O, Dash C, Okeoma CM. Effect of prolonged freezing of semen on exosome recovery and biologic activity. Sci Rep. 2017;7:45034. doi: 10.1038/srep45034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Menezes EGM, Jang K, George AF, Nyegaard M, Neidleman J, Inglis HC, Danesh A, Deng X, Afshari A, Kim YH. Seminal plasma-derived extracellular-vesicle fractions from HIV-infected men exhibit unique microRNA signatures and induce a proinflammatory response in cells isolated from the female reproductive tract. J Virol. 2020;94:e0525. doi: 10.1128/JVI.00525-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouattara LA, Anderson SM, Doncel GF. Seminal exosomes and HIV-1 transmission. Andrologia. 2018;50:e13220. doi: 10.1111/and.13220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paktinat S, Hashemi SM, Novin MG, Mohammadi-Yeganeh S, Salehpour S, Karamian A, Nazarian H. Seminal exosomes induce interleukin-6 and interleukin-8 secretion by human endometrial stromal cells. Eur J Obstet Gynecol Reprod Biol. 2019;235:71–76. doi: 10.1016/j.ejogrb.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Simon C, Greening DW, Bolumar D, Balaguer N, Salamonsen LA, Vilella F. Extracellular vesicles in human reproduction in health and disease. Endocr Rev. 2018;39:292–332. doi: 10.1210/er.2017-00229. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan R, Saez F. Epididymosomes, prostasomes, and liposomes: their roles in mammalian male reproductive physiology. Reproduction. 2013;146:R21–R35. doi: 10.1530/REP-13-0058. [DOI] [PubMed] [Google Scholar]
- 15.Mimiaga MJ, Reisner SL, Grasso C, Crane HM, Safren SA, Kitahata MM, Schumacher JE, Mathews WC, Mayer KH. Substance use among HIV-infected patients engaged in primary care in the United States: findings from the Centers for AIDS Research Network of Integrated Clinical Systems cohort. Am J Public Health. 2013;103:1457–1467. doi: 10.2105/AJPH.2012.301162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohammadi A, Darabi M, Nasry M, Saabet-Jahromi MJ, Malek-Pour-Afshar R, Sheibani H. Effect of opium addiction on lipid profile and atherosclerosis formation in hypercholesterolemic rabbits. Exp Toxicol Pathol. 2009;61:145–149. doi: 10.1016/j.etp.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Roohafza H, Talaei M, Sadeghi M, Haghani P, Shokouh P, Sarrafzadegan N. Opium decreases the age at myocardial infarction and sudden cardiac death: a long- and short-term outcome evaluation. Arch Iran Med. 2013;16:154–160. [PubMed] [Google Scholar]
- 18.Nabati S, Asadikaram G, Arababadi MK, Shahabinejad G, Rezaeian M, Mahmoodi M, Kennedy D. The plasma levels of the cytokines in opium-addicts and the effects of opium on the cytokines secretion by their lymphocytes. Immunol Lett. 2013;152:42–46. doi: 10.1016/j.imlet.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Saha B, Momen-Heravi F, Kodys K, Szabo G. MicroRNA cargo of extracellular vesicles from alcohol-exposed monocytes signals naive monocytes to differentiate into M2 macrophages. J Biol Chem. 2016;291:149–159. doi: 10.1074/jbc.M115.694133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Momen-Heravi F, Saha B, Kodys K, Catalano D, Satishchandran A, Szabo G. Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J Transl Med. 2015;13:261. doi: 10.1186/s12967-015-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Momen-Heravi F, Bala S, Kodys K, Szabo G. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci Rep. 2015;5:9991. doi: 10.1038/srep09991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fronczak CM, Kim ED, Barqawi AB. The insults of illicit drug use on male fertility. J Androl. 2012;33:515–528. doi: 10.2164/jandrol.110.011874. [DOI] [PubMed] [Google Scholar]
- 23.Krieger JN, Coombs RW, Collier AC, Koehler JK, Ross SO, Chaloupka K, Murphy VL, Corey L. Fertility parameters in men infected with human immunodeficiency virus. J Infect Dis. 1991;164:464–469. doi: 10.1093/infdis/164.3.464. [DOI] [PubMed] [Google Scholar]
- 24.Nicopoullos J, Almeida P, Vourliotis M, Gilling-Smith C. A decade of the sperm-washing programme: correlation between markers of HIV and seminal parameters. HIV Med. 2011;12:195–201. doi: 10.1111/j.1468-1293.2010.00868.x. [DOI] [PubMed] [Google Scholar]
- 25.Dulioust E, Du AL, Costagliola D, Guibert J, Kunstmann J-M, Heard I, Juillard J-C, Salmon D, Leruez-Ville M, Mandelbrot L. Semen alterations in HIV-1 infected men. Hum Reprod. 2002;17:2112–2118. doi: 10.1093/humrep/17.8.2112. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Ho W-Z. Drugs of abuse and HIV infection/replication: implications for mother–fetus transmission. Life Sci. 2011;88:972–979. doi: 10.1016/j.lfs.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapadia F, Vlahov D, Donahoe RM, Friedland G. The role of substance abuse in HIV disease progression: reconciling differences from laboratory and epidemiologic investigations. Clin Infect Dis. 2005;41:1027–1034. doi: 10.1086/433175. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Wang J, Zhang H-R, Shi H-J, Ma D, Zhao H-X, Lin B, Li R-S. Proteomic analysis of seminal plasma from asthenozoospermia patients reveals proteins that affect oxidative stress responses and semen quality. Asian J Androl. 2009;11:484. doi: 10.1038/aja.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batruch I, Smith CR, Mullen BJ, Grober E, Lo KC, Diamandis EP, Jarvi KA. Analysis of seminal plasma from patients with non-obstructive azoospermia and identification of Candidate Biomarkers of Male Infertility. J Proteome Res. 2012;11:1503–1511. doi: 10.1021/pr200812p. [DOI] [PubMed] [Google Scholar]
- 30.Drabovich AP, Jarvi K, Diamandis EP. Verification of male infertility biomarkers in seminal plasma by multiplex selected reaction monitoring assay. Mol Cell Prot. 2011;10:e004127. doi: 10.1074/mcp.M110.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu W, Hu Z, Qin Y, Dong J, Dai J, Lu C, Zhang W, Shen H, Xia Y, Wang X. Seminal plasma microRNAs: potential biomarkers for spermatogenesis status. Mol Hum Reprod. 2012;18:489–497. doi: 10.1093/molehr/gas022. [DOI] [PubMed] [Google Scholar]
- 32.Luo L-Y, Shan SJ, Elliott MB, Soosaipillai A, Diamandis EP. Purification and characterization of human kallikrein 11, a candidate prostate and ovarian cancer biomarker, from seminal plasma. Clin Cancer Res. 2006;12:742–750. doi: 10.1158/1078-0432.CCR-05-1696. [DOI] [PubMed] [Google Scholar]
- 33.Drabovich AP, Saraon P, Jarvi K, Diamandis EP. Seminal plasma as a diagnostic fluid for male reproductive system disorders. Nat Rev Urol. 2014;11:278. doi: 10.1038/nrurol.2014.74. [DOI] [PubMed] [Google Scholar]
- 34.Pelloni M, Coltrinari G, Paoli D, Pallotti F, Lombardo F, Lenzi A, Gandini L. Differential expression of miRNAs in the seminal plasma and serum of testicular cancer patients. Endocrine. 2017;57:518–527. doi: 10.1007/s12020-016-1150-z. [DOI] [PubMed] [Google Scholar]
- 35.Hong Y, Wang C, Fu Z, Liang H, Zhang S, Lu M, Sun W, Ye C, Zhang C-Y, Zen K. Systematic characterization of seminal plasma piRNAs as molecular biomarkers for male infertility. Sci Rep. 2016;6:24229. doi: 10.1038/srep24229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barceló M, Castells M, Bassas L, Vigués F, Larriba S. Semen miRNAs contained in exosomes as non-invasive biomarkers for prostate cancer diagnosis. Sci Rep. 2019;9:13772. doi: 10.1038/s41598-019-50172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Alessandro A, Zolla L. Meat science: from proteomics to integrated omics towards system biology. J Proteomics. 2013;78:558–577. doi: 10.1016/j.jprot.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 38.Fukushima A, Kusano M, Redestig H, Arita M, Saito K. Integrated omics approaches in plant systems biology. Curr Opin Chem Biol. 2009;13:532–538. doi: 10.1016/j.cbpa.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 39.Misra BB, Langefeld C, Olivier M, Cox LA. Integrated omics: tools, advances and future approaches. J Mol Endocrinol. 2019;62:R21–R45. doi: 10.1530/JME-18-0055. [DOI] [PubMed] [Google Scholar]
- 40.Lyu Y, Kaddour H, Kopcho S, Panzner TD, Shouman N, Kim E-Y, Martinson J, McKay H, Martinez-Maza O, Margolick JB, et al. Human immunodeficiency virus (HIV) infection and use of illicit substances promote secretion of semen exosomes that enhance monocyte adhesion and induce actin reorganization and chemotactic migration. Cells. 2019;8:1027. doi: 10.3390/cells8091027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seidler S, Zimmermann HW, Bartneck M, Trautwein C, Tacke F. Age-dependent alterations of monocyte subsets and monocyte-related chemokine pathways in healthy adults. BMC Immunol. 2010;11:30. doi: 10.1186/1471-2172-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Martinis M, Modesti M, Ginaldi L. Phenotypic and functional changes of circulating monocytes and polymorphonuclear leucocytes from elderly persons. Immunol Cell Biol. 2004;82:415–420. doi: 10.1111/j.0818-9641.2004.01242.x. [DOI] [PubMed] [Google Scholar]
- 43.Gerhardt T, Ley K. Monocyte trafficking across the vessel wall. Cardiovasc Res. 2015;107:321–330. doi: 10.1093/cvr/cvv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witt CM, Raychaudhuri S, Schaefer B, Chakraborty AK, Robey EA. Directed migration of positively selected thymocytes visualized in real time. PLoS Biol. 2005;3:e160. doi: 10.1371/journal.pbio.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong CH, Heit B, Kubes P. Molecular regulators of leucocyte chemotaxis during inflammation. Cardiovasc Res. 2010;86:183–191. doi: 10.1093/cvr/cvq040. [DOI] [PubMed] [Google Scholar]
- 46.Moser B, Loetscher P. Lymphocyte traffic control by chemokines. Nat Immunol. 2001;2:123–128. doi: 10.1038/84219. [DOI] [PubMed] [Google Scholar]
- 47.Sánchez-Madrid F, del Pozo MA. Leukocyte polarization in cell migration and immune interactions. EMBO J. 1999;18:501–511. doi: 10.1093/emboj/18.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loetscher P, Moser B, Baggiolini M. Chemokines and their receptors in lymphocyte traffic and HIV infection. Adv Immunol. 1999;74:127–180. doi: 10.1016/s0065-2776(08)60910-4. [DOI] [PubMed] [Google Scholar]
- 49.Paskauskas S, Parseliunas A, Kerkadze V, Nobiling R, Schmidt J, Ryschich E. Blockade of leukocyte haptokinesis and haptotaxis by ketoprofen, diclofenac and SC-560. BMC Immunol. 2011;12:1–9. doi: 10.1186/1471-2172-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weninger W, Biro M, Jain R. Leukocyte migration in the interstitial space of non-lymphoid organs. Nat Rev Immunol. 2014;14:232–246. doi: 10.1038/nri3641. [DOI] [PubMed] [Google Scholar]
- 51.Ganiko L, Martins AR, Freymüller E, Mortara RA, Roque-Barreira M-C. Lectin KM+-induced neutrophil haptotaxis involves binding to laminin. Biochim Biophys Acta. 2005;1721:152–163. doi: 10.1016/j.bbagen.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 52.Kaslow RA, Ostrow DG, Detels R, Phair JP, Polk BF. RINALDO Jr CR, Study MAC: The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am J Epidemiol. 1987;126:310–318. doi: 10.1093/aje/126.2.310. [DOI] [PubMed] [Google Scholar]
- 53.Detels R, Jacobson L, Margolick J, Martinez-Maza O, Muñoz A, Phair J, Rinaldo C, Wolinsky S. The multicenter AIDS cohort study, 1983 to…. Public Health. 2012;126:196–198. doi: 10.1016/j.puhe.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaddour H, Panzner TD, Welch JL, Shouman N, Mohan M, Stapleton JT, Okeoma CM. Electrostatic surface properties of blood and semen extracellular vesicles: implications of sialylation and HIV-induced changes on EV internalization. Viruses. 2020;12:1117. doi: 10.3390/v12101117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 56.Real R, Vargas JM. The probabilistic basis of Jaccard’s index of similarity. Syst Biol. 1996;45:380–385. [Google Scholar]
- 57.Yang C, Guo WB, Zhang WS, Bian J, Yang JK, Zhou QZ, Chen MK, Peng W, Qi T, Wang CY. Comprehensive proteomics analysis of exosomes derived from human seminal plasma. Andrology. 2017;5:1007–1015. doi: 10.1111/andr.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murdica V, Cermisoni GC, Zarovni N, Salonia A, Viganò P, Vago R. Proteomic analysis reveals the negative modulator of sperm function glycodelin as over-represented in semen exosomes isolated from asthenozoospermic patients. Hum Reprod. 2019;34:1416–1427. doi: 10.1093/humrep/dez114. [DOI] [PubMed] [Google Scholar]
- 59.Lin Y, Liang A, He Y, Li Z, Li Z, Wang G, Sun F. Proteomic analysis of seminal extracellular vesicle proteins involved in asthenozoospermia by iTRAQ. Mol Reprod Dev. 2019;86:1094–1105. doi: 10.1002/mrd.23224. [DOI] [PubMed] [Google Scholar]
- 60.Kaddour H, Lyu Y, Welch JL, Paromov V, Mandape SN, Sakhare SS, Pandhare J, Stapleton JT, Pratap S, Dash C. Proteomics profiling of autologous blood and semen exosomes from HIV-infected and uninfected individuals reveals compositional and functional variabilities. Mol Cell Proteomics. 2020;19:78–100. doi: 10.1074/mcp.RA119.001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barberán-Soler S, Vo JM, Hogans RE, Dallas A, Johnston BH, Kazakov SA. Decreasing miRNA sequencing bias using a single adapter and circularization approach. Genome Biol. 2018;19:1–9. doi: 10.1186/s13059-018-1488-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chung IF, Chang S-J, Chen C-Y, Liu S-H, Li C-Y, Chan C-H, Shih C-C, Cheng W-C. YM500v3: a database for small RNA sequencing in human cancer research. Nucleic Acids Res. 2017;45:D925–D931. doi: 10.1093/nar/gkw1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Persengiev SP, Kondova II, Bontrop RE. The Impact of MicroRNAs on Brain Aging and Neurodegeneration. Curr Gerontol Geriatr Res. 2012;2012:359369. doi: 10.1155/2012/359369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. NeuroReport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 67.Wu L, Zhang G, Guo C, Zhao X, Shen D, Yang N. MiR-128-3p mediates TNF-α-induced inflammatory responses by regulating Sirt1 expression in bone marrow mesenchymal stem cells. Biochem Biophys Res Commun. 2020;521:98–105. doi: 10.1016/j.bbrc.2019.10.083. [DOI] [PubMed] [Google Scholar]
- 68.Li J, Zhu L, Su H, Liu D, Yan Z, Ni T, Wei H, Goh ELK, Chen T. Regulation of miR-128 in the nucleus accumbens affects methamphetamine-induced behavioral sensitization by modulating proteins involved in neuroplasticity. Addict Biol. 2020;26:12881. doi: 10.1111/adb.12881. [DOI] [PubMed] [Google Scholar]
- 69.Zhang T, Yu J, Zhang Y, Li L, Chen Y, Li D, Liu F, Zhang CY, Gu H, Zen K. Salmonella enterica serovar enteritidis modulates intestinal epithelial miR-128 levels to decrease macrophage recruitment via macrophage colony-stimulating factor. J Infect Dis. 2014;209:2000–2011. doi: 10.1093/infdis/jiu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eletto D, Russo G, Passiatore G, Del Valle L, Giordano A, Khalili K, Gualco E, Peruzzi F. Inhibition of SNAP25 expression by HIV-1 Tat involves the activity of mir-128a. J Cell Physiol. 2008;216:764–770. doi: 10.1002/jcp.21452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carossino M, Dini P, Kalbfleisch TS, Loynachan AT, Canisso IF, Shuck KM, Timoney PJ, Cook RF, Balasuriya UBR. Downregulation of microRNA eca-mir-128 in seminal exosomes and enhanced expression of CXCL16 in the stallion reproductive tract are Associated with long-term persistence of equine arteritis virus. J Virol. 2018;92:e00015. doi: 10.1128/JVI.00015-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bondanese VP, Francisco-Garcia A, Bedke N, Davies DE, Sanchez-Elsner T. Identification of host miRNAs that may limit human rhinovirus replication. World J Biol Chem. 2014;5:437–456. doi: 10.4331/wjbc.v5.i4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cai J, Fang L, Huang Y, Li R, Xu X, Hu Z, Zhang L, Yang Y, Zhu X, Zhang H, et al. Simultaneous overactivation of Wnt/β-catenin and TGFβ signalling by miR-128-3p confers chemoresistance-associated metastasis in NSCLC. Nat Commun. 2017;8:15870. doi: 10.1038/ncomms15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Franzoni E, Booker SA, Parthasarathy S, Rehfeld F, Grosser S, Srivatsa S, Fuchs HR, Tarabykin V, Vida I, Wulczyn FG. miR-128 regulates neuronal migration, outgrowth and intrinsic excitability via the intellectual disability gene Phf6. Elife. 2015 doi: 10.7554/eLife.04263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, Wang L, Li P, Zhao Y, Duan W, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18:43. doi: 10.1186/s12943-019-0981-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Pan J, Zhou C, Zhao X, He J, Tian H, Shen W, Han Y, Chen J, Fang S, Meng X, et al. A two-miRNA signature (miR-33a-5p and miR-128-3p) in whole blood as potential biomarker for early diagnosis of lung cancer. Sci Rep. 2018;8:16699. doi: 10.1038/s41598-018-35139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang L, Zhang L, Lu L, Wang Y. Long noncoding RNA SNHG16 sponges miR-182-5p and miR-128-3p to promote retinoblastoma cell migration and invasion by targeting LASP1. Onco Targets Ther. 2019;12:8653–8662. doi: 10.2147/OTT.S212352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao L, Li R, Xu S, Li Y, Zhao P, Dong W, Liu Z, Zhao Q, Tan B. Tumor suppressor miR-128-3p inhibits metastasis and epithelial-mesenchymal transition by targeting ZEB1 in esophageal squamous-cell cancer. Acta Biochim Biophys Sin (Shanghai) 2018;50:171–180. doi: 10.1093/abbs/gmx132. [DOI] [PubMed] [Google Scholar]
- 79.Zhou T, Wu L, Wang Q, Jiang Z, Li Y, Ma N, Chen W, Hou Z, Gan W, Chen S. MicroRNA-128 targeting RPN2 inhibits cell proliferation and migration through the Akt-p53-cyclin pathway in colorectal cancer cells. Oncol Lett. 2018;16:6940–6949. doi: 10.3892/ol.2018.9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou XU, Qi L, Tong S, Cui YU, Chen J, Huang T, Chen Z, Zu XB. miR-128 downregulation promotes growth and metastasis of bladder cancer cells and involves VEGF-C upregulation. Oncol Lett. 2015;10:3183–3190. doi: 10.3892/ol.2015.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kong D, Zhang Z. NAIF1 suppresses osteosarcoma progression and is regulated by miR-128. Cell Biochem Funct. 2018;36:443–449. doi: 10.1002/cbf.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.She X, Yu Z, Cui Y, Lei Q, Wang Z, Xu G, Xiang J, Wu M, Li G. miR-128 and miR-149 enhance the chemosensitivity of temozolomide by Rap1B-mediated cytoskeletal remodeling in glioblastoma. Oncol Rep. 2014;32:957–964. doi: 10.3892/or.2014.3318. [DOI] [PubMed] [Google Scholar]
- 83.Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS ONE. 2018;13:e0206239. doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mou T, Luo Y, Huang Z, Zheng D, Pu X, Shen A, Pu J, Li T, Dai J, Chen W, Wu Z. Inhibition of microRNA-128-3p alleviates liver ischaemia-reperfusion injury in mice through repressing the Rnd3/NF-κB axis. Innate Immun. 2020;26:528–536. doi: 10.1177/1753425920928449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chandra LC, Kumar V, Torben W, Vande Stouwe C, Winsauer P, Amedee A, Molina PE, Mohan M. Chronic administration of Delta9-tetrahydrocannabinol induces intestinal anti-inflammatory microRNA expression during acute simian immunodeficiency virus infection of rhesus macaques. J Virol. 2015;89:1168–1181. doi: 10.1128/JVI.01754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kumar V, Torben W, Mansfield J, Alvarez X, Vande Stouwe C, Li J, Byrareddy SN, Didier PJ, Pahar B, Molina PE, Mohan M. Cannabinoid attenuation of intestinal inflammation in chronic SIV-infected rhesus macaques involves T Cell modulation and differential expression of micro-RNAs and pro-inflammatory genes. Front Immunol. 2019;10:914. doi: 10.3389/fimmu.2019.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 88.Balgley BM, Laudeman T, Yang L, Song T, Lee CS. Comparative evaluation of tandem MS search algorithms using a target-decoy search strategy. Mol Cell Proteomics. 2007;6:1599–1608. doi: 10.1074/mcp.M600469-MCP200. [DOI] [PubMed] [Google Scholar]
- 89.Fang X, Balgley BM, Wang W, Park DM, Lee CS. Comparison of multidimensional shotgun technologies targeting tissue proteomics. Electrophoresis. 2009;30:4063–4070. doi: 10.1002/elps.200900367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schenkel AR, Dufour EM, Chew TW, Sorg E, Muller WA. The murine CD99-related molecule CD99-Like 2 (CD99L2) is an adhesion molecule involved in the inflammatory response. Cell Commun Adhes. 2007;14:227–237. doi: 10.1080/15419060701755966. [DOI] [PubMed] [Google Scholar]
- 91.Bixel MG, Li H, Petri B, Khandoga AG, Khandoga A, Zarbock A, Wolburg-Buchholz K, Wolburg H, Sorokin L, Zeuschner D, et al. CD99 and CD99L2 act at the same site as, but independently of, PECAM-1 during leukocyte diapedesis. Blood. 2010;116:1172–1184. doi: 10.1182/blood-2009-12-256388. [DOI] [PubMed] [Google Scholar]
- 92.Rakhilin SV, Olson PA, Nishi A, Starkova NN, Fienberg AA, Nairn AC, Surmeier DJ, Greengard P. A network of control mediated by regulator of calcium/calmodulin-dependent signaling. Science. 2004;306:698–701. doi: 10.1126/science.1099961. [DOI] [PubMed] [Google Scholar]
- 93.Smirnova L, Gräfe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 94.Zou L, Cheng G, Xu C, Liu H, Wang Y, Li N, Zhu C, Xia W. The role of miR-128-3p through MAPK14 activation in the apoptosis of GC2 spermatocyte cell line following heat stress. Andrology. 2021;9:665–672. doi: 10.1111/andr.12923. [DOI] [PubMed] [Google Scholar]
- 95.Carossino M, Dini P, Kalbfleisch TS, Loynachan AT, Canisso IF, Shuck KM, Timoney PJ, Cook RF, Balasuriya UB. Downregulation of microRNA eca-mir-128 in seminal exosomes and enhanced expression of CXCL16 in the stallion reproductive tract are associated with long-term persistence of equine arteritis virus. J Virol. 2018 doi: 10.1128/JVI.00015-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bochnakian A, Zhen A, Zisoulis DG, Idica A, KewalRamani VN, Neel N, Daugaard I, Hamdorf M, Kitchen S, Lee K, Pedersen IM. Interferon-inducible MicroRNA miR-128 modulates HIV-1 replication by targeting TNPO3 mRNA. J Virol. 2019 doi: 10.1128/JVI.00364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Petrulis A. Chemosignals and hormones in the neural control of mammalian sexual behavior. Front Neuroendocrinol. 2013;34:255–267. doi: 10.1016/j.yfrne.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 98.Li J, Xie M, Wang X, Ouyang X, Wan Y, Dong G, Yang Z, Yang J, Yue J. Sex hormones regulate cerebral drug metabolism via brain miRNAs: down-regulation of brain CYP2D by androgens reduces the analgesic effects of tramadol. Br J Pharmacol. 2015;172:4639–4654. doi: 10.1111/bph.13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li H, Li C, Zhou Y, Luo C, Ou J, Li J, Mo Z. Expression of microRNAs in the serum exosomes of methamphetamine-dependent rats vs. ketamine-dependent rats. Exp Ther Med. 2018;15:3369–3375. doi: 10.3892/etm.2018.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li HC, Lin YB, Li C, Luo CH, Zhou YT, Ou JY, Li J, Mo ZX. Expression of miRNAs in Serum Exosomes versus Hippocampus in Methamphetamine-induced rats and intervention of rhynchophylline. Evid Based Complement Alternat Med. 2018;2018:8025062. doi: 10.1155/2018/8025062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sandau US, Duggan E, Shi X, Smith SJ, Huckans M, Schutzer WE, Loftis JM, Janowsky A, Nolan JP, Saugstad JA. Methamphetamine use alters human plasma extracellular vesicles and their microRNA cargo: an exploratory study. J Extracell Vesicles. 2020;10:e12028. doi: 10.1002/jev2.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Evangelisti C, Florian MC, Massimi I, Dominici C, Giannini G, Galardi S, Buè MC, Massalini S, McDowell HP, Messi E, et al. MiR-128 up-regulation inhibits Reelin and DCX expression and reduces neuroblastoma cell motility and invasiveness. Faseb J. 2009;23:4276–4287. doi: 10.1096/fj.09-134965. [DOI] [PubMed] [Google Scholar]
- 103.Wang R, Gornalusse GG, Kim Y, Pandey U, Hladik F, Vojtech L. Potent restriction of sexual Zika Virus infection by the lipid fraction of extracellular vesicles in semen. Front Microbiol. 2020;11:574054. doi: 10.3389/fmicb.2020.574054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maciel E, Mansuy IM. Extracellular vesicles and their miRNA Cargo: a means of communication between soma and germline in the mammalian reproductive system. Chimia (Aarau) 2019;73:356–361. doi: 10.2533/chimia.2019.356. [DOI] [PubMed] [Google Scholar]
- 105.Chan JC, Morgan CP, Adrian Leu N, Shetty A, Cisse YM, Nugent BM, Morrison KE, Jašarević E, Huang W, Kanyuch N, et al. Reproductive tract extracellular vesicles are sufficient to transmit intergenerational stress and program neurodevelopment. Nat Commun. 2020;11:1499. doi: 10.1038/s41467-020-15305-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 107.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–d302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data sets supporting the conclusions of this article have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD020487. The sRNA-Seq data sets are included within the article and its additional files.