Abstract
Alcohol‐associated liver disease (ALD) is a major cause of alcohol‐related mortality. Sex differences in sensitivity to ALD are well described, but these are often disregarded in studies of ALD development. We aimed to define sex‐specific pathways in liver exposed to alcohol. Mice were fed the Lieber‐DeCarli alcohol liquid diet or a combination of a high‐fat diet with alcohol in water. Single‐cell RNA sequencing (scRNA‐Seq) was performed on liver cells from male and female mice. Mice were treated with adeno‐associated virus (AAV)‐short hairpin (sh)Control or AAV‐sh lysine demethylase 5b (shKdm5b) and/or AAV‐shKdm5c vectors. Changes after Kdm5b/5c knockdown were assessed by RNA‐Seq and histone H3 lysine K4 (H3K4)me3 chromatin immunoprecipitation‐Seq analysis. Using scRNA‐Seq analysis, we found several sex‐specific pathways induced by alcohol, including pathways related to lipid metabolism and hepatocyte differentiation. Bioinformatic analysis suggested that two epigenetic regulators, H3K4‐specific lysine demethylases KDM5B and KDM5C, contribute to sex differences in alcohol effects. We found that in alcohol‐fed male mice, KDM5B and KDM5C are involved in hepatocyte nuclear factor 4 alpha (Hnf4a) down‐regulation, hepatocyte dedifferentiation, and an increase in fatty acid synthesis. This effect is mediated by alcohol‐induced KDM5B and KDM5C recruitment to Hnf4a and other gene promoters in male but not in female mice. Kdm5b and Kdm5c knockdown or KDM5‐inhibitor treatment prevented alcohol‐induced lipid accumulation and restored levels of Hnf4a and other hepatocyte differentiation genes in male mice. In addition, Kdm5b knockdown prevented hepatocellular carcinoma development in male mice by up‐regulating Hnf4a and decreasing tumor cell proliferation. Conclusion: Alcohol specifically activates KDM5 demethylases in male mice to promote alcohol‐induced hepatocyte dedifferentiation and tumor development.
Abbreviations
- AAV
adeno‐associated virus
- AH
alcohol‐associated hepatitis
- Alb
albumin
- ALD
alcohol‐associated liver disease
- ALT
alanine aminotransferase
- AR
androgen receptor
- BSA
bovine serum albumin
- ChIP
chromatin immunoprecipitation
- Cyp2e1
cytochrome P450 family 2 subfamily E member 1
- EDTA
ethylene diamine tetraacetic acid
- Fasn
fatty acid synthase
- GO
Gene Ontology
- H&E
hematoxylin and eosin
- H3K4
histone H3 lysine K4
- HCC
hepatocellular carcinoma
- HNF4
hepatocyte nuclear factor 4
- HSP
heat shock protein
- KDM5B/5C
lysine demethylase 5B/5C
- KUMC
University of Kansas Medical Center
- mRNA
messenger RNA
- PBS
phosphate‐buffered saline
- Ppara
peroxisome proliferator‐activated receptor alpha
- PT
prothrombin time
- ScdI
stearoyl‐coenzyme A desaturase 1
- scrmb
scramble
- scRNA
single‐cell RNA
- Seq
sequencing
- shRNA
short hairpin RNA
- TG
triglyceride
- Tris
trishydroxymethylaminomethane
- WD
Western diet
- WDA
Western diet alcohol
Alcohol‐associated liver disease (ALD) is a complex disease with a variable phenotype that is affected by a combination of genetic and environmental factors. Sex is an important variable, and it has been recognized for decades that the consequences of alcohol consumption are different between the sexes.( 1 , 2 , 3 , 4 , 5 ) Female sex is an independent predictor of mortality in acute alcohol‐associated hepatitis (AH), although men with AH have lower median platelet counts and higher serum creatinine, alanine aminotransferase (ALT), and gamma‐glutamytransferase concentrations.( 1 ) Several studies reported that female animals have a more pronounced inflammatory response to alcohol, both in adults and in alcohol‐exposed embryos, and the alcohol‐stimulated accumulation of chemokine (C‐C motif) ligand 2 in the hypothalamus of embryos and neonates is sexually dimorphic.( 2 )
Other studies indicate that alcohol metabolism is different between the sexes. Hepatic alcohol dehydrogenase activity, for example, is higher in female than in male rats,( 5 ) and this sexually dimorphic expression pattern is mediated by sex‐specific growth hormone secretion. In addition, differences between male and female cells in sensitivity to ethanol persist independently of endogenous hormones.( 4 ) Sex differences in expression of cytochrome P450 family 2 subfamily E member 1 (CYP2E1) have been reported to result from epigenetic silencing of Cyp2e1 in male mice,( 4 ) and sex‐specific differences in expression of aldehyde dehydrogenase 1 family member B1 (ALDH1B1) contribute to sex differences in susceptibility to hepatic steatosis and disease progression.( 6 )
Additional studies have noted that, apart from differential expression of alcohol‐metabolizing enzymes, alcohol induced higher expression of heat shock protein 27 (HSP27) and HSP70, higher levels of hepatocyte proliferation, and interleukin‐6 signaling pathway activation in male mice, while in female mice, alcohol induced more hepatocyte apoptosis.( 7 )
Lysine demethylase 5B (KDM5B) and KDM5C are histone demethylases that regulate the methylation state of histone H3 lysine K4 (H3K4) and play a central role in epigenetic regulation. The H3K4 demethylases remove methyl groups from lysine 4, leading to loss of RNA polymerase binding and transcriptional repression. These demethylases were first identified as involved in brain development, and they have been reported to be mutated in both autism and Rett syndrome.( 8 ) In addition, KDM5 demethylases are involved in cancer development,( 9 , 10 , 11 , 12 , 13 ) inflammation control,( 14 ) and replication stress responses.( 15 ) Several studies indicate that KDM5B binds androgen receptor (AR) and is involved in prostate cancer pathogenesis, although the role of KDM5B in prostate development and prostate cancer is likely context dependent.( 9 , 10 , 11 , 16 ) Thus, there is an indication for sex‐specific roles of KDM5B.
In this work, we have shown that alcohol induces differential gene expression responses in male and female liver cells. We identified several key pathways, including regeneration, proliferation, inflammation, and xenobiotic and lipid metabolism, that are differentially regulated in male and female mice in response to alcohol and found several epigenetic regulators associated with these sex differences. These epigenetic regulators include the histone lysine demethylases KDM5B and KDM5C, which are specific to histone H3K4 methylation. We found that in alcohol‐fed mice, KDM5B and KDM5C mediate multiple sex‐specific changes, including hepatocyte nuclear factor 4 alpha (Hnf4a) down‐regulation, suppression of clotting factor synthesis, promotion of liver injury, and alteration of lipid synthesis. Taken together, these results show that KDM5 demethylases are regulators of sex‐specific mechanisms in the hepatic response to alcohol.
Materials and Methods
Mice and Feeding Procedures
We purchased 6‐7‐week‐old C57BL6/J mice from Jackson Laboratory. All mice were housed in a temperature‐controlled, specific, pathogen‐free environment with 12‐hour light–dark cycles. All animal‐handling procedures were approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center (KUMC) (Kansas City, KS).
Lieber‐DeCarli liquid diet feeding was performed as described.( 17 ) Both male and female mice received control or alcohol liquid diet (4.8% alcohol) for 3‐5 weeks. For the previously described Western diet alcohol (WDA) model,( 18 ) both male and female mice were fed ad libitum Western diet (WD) (Research Diets, Inc., Cat. No. D12079B; 40% calories from fat [90% milk fat, 10% corn oil], 0.2% cholesterol), and alcohol was given ad libitum in water at indicated concentrations. Mice in alcohol groups received a progressively increasing amount of alcohol in water (1%, 3%, 10%, 15%, and 20% for 3 days each). After reaching 20%, mice were then alternated between 20% (4 days, Thursday until Monday) and 10% (3 days, Monday until Thursday).
Vectors
pUB_smFLAG_KDM5B was provided by Tim Stasevich through Addgene (Addgene plasmid 81084).
Adeno‐associated virus (AAV)8‐U6‐m‐aryl hydrocarbon receptor (AhR)‐short hairpin (sh)RNA, AAV8‐green fluorescent protein (GFP)‐U6‐scramble (scrmb)‐shRNA, AAV9‐U6‐m‐Kdm5b‐shRNA, AAV9‐U6‐m‐Kdm5c‐shRNA, and AAV9‐U6‐scrmb‐shRNA were from Vector Biolabs, Malvern, PA.
Human and mouse shRNA vectors were from Sigma (Cat. No. TRCN0000014759, TRCN0000379331, TRCN0000295348, TRCN0000022087).
Antibodies
Anti‐Hnf4α antibodies were from Novus. Anti‐KDM5B, anti‐KDM5C, and anti‐H3K4me3 antibodies were from Cell Signaling. Anti‐β‐actin was from Santa Cruz.
Liver Cell Isolation
Liver cells were isolated by a modification of the method described by Troutman et al.( 19 ) Mouse livers were digested by retrograde perfusion with liberase through the inferior vena cava. The dissociated cell mixture was placed into a 50‐mL conical tube and centrifuged twice at 50g for 2 minutes to pellet hepatocytes.
Single‐Cell RNA Sequencing
Live hepatocytes and neural progenitor cells were purified using the dead‐cell removal kit (MiltenyiBiotec). Liver cells were immediately used to generate barcoded complementary DNA (cDNA) libraries, using a 10× Genomics Chromium platform with a total input of 10,000 cells per condition. Libraries were sequenced with an Illumina Novoseq sequencer, and data were analyzed with the 10× Genomics Cell Ranger and Loupe Cell Browser software.
RNA Sequencing
For RNA sequencing (RNA‐Seq) analysis, total RNA was isolated from liver by using the Qiagen RNA isolation kit. Three individual mice per condition were used. Library generation and sequencing was performed by BGI genomics services (Cambridge, MA). Twenty‐four samples were sequenced using the BGISEQ platform, on average generating about 4.57 gigabyte bases per sample. The average mapping ratio with a reference genome was 96.14%; 16,869 genes were identified. Differential gene expression was identified with DESeq2.
Chromatin Immunoprecipitation‐SEQ
Chromatin immunoprecipitation (ChIP)‐Seq was performed by Active Motif (Carlsbad, CA), using H3K4me3 antibodies (Cat. No. 39159) at 4 µL per chromatin sample. Peaks were called using the MACS 2.1.0 algorithm. MACS cutoff was P = 1 × 10−7 for narrow peaks and P = 1 × 10−1 for broad peaks. Peak filtering was performed by removing false ChIP‐Seq peaks as defined within the Encyclopedia of DNA Elements blacklist. Top differential regions were identified using DESeq2 (Supporting Table S2). A differential region motif analysis was performed using HOMER.
ChIP
ChIP was performed as described.( 20 , 21 ) Whole‐liver cells were crosslinked by the addition of 1% formaldehyde for 10 minutes. Cells were lysed with 10 mM trishydroxymethylaminomethane (Tris)‐HCl (pH 8.0), 10 mM NaCl, 3 mM MgCl2, and 0.5% Nonidet P40 (NP‐40). Nuclei were collected by centrifugation; resuspended in 1% sodium dodecyl sulfate (SDS), 5 mmol/L ethylene diamine tetraacetic acid (EDTA), and 50 mmol/L Tris‐HCl (pH 8.0); and sonicated to generate chromatin to an average length of ~100 to 500 base pairs. Next, samples in 1% Triton X‐100, 2 mM EDTA, 20 mM Tris‐HCl (pH 8.1), and 150 mM NaCl were immunoprecipitated overnight at 4°C with 4 μg ChIP‐grade antibody. We used 20 µL of magnetic beads (Dynabeads M‐280; Invitrogen) to purify immunocomplexes. Following purification, crosslinks were reverted by incubation at 65°C for 6 hours. Samples were purified with the Qiagen DNA purification kit.
Western Blot
Protein extracts (15 µg) were subjected to 10% SDS‐polyacrylamide gel electrophoresis, electrophoretically transferred to nitrocellulose membranes (Amersham Hybond enhanced chemiluminescence [ECL]; GE Healthcare), and blocked in 3% bovine serum albumin (BSA)/phosphate‐buffered saline (PBS) at room temperature for 1 hour. Primary antibodies were incubated overnight at manufacturer‐recommended concentrations. Immunoblots were detected with the ECL Plus Western Blotting Detection System (Amersham Biosciences, Piscataway, NJ) or using near‐infrared fluorescence with the ODYSSEY Fc, Dual‐Mode Imaging system (Li‐COR).
Immunohistochemistry
Immunostaining on formalin‐fixed sections was performed by deparaffinization and rehydration followed by antigen retrieval by heating in a pressure cooker (121°C) for 5 minutes in 10 mM sodium citrate (pH 6.0) as described.( 22 ) Sections were rinsed 3 times in PBS/PBS‐0.1% Tween‐20 and incubated in Dako Protein Block (Dako, Carpinteria, CA) at room temperature for 1 hour. After removal of the blocking solution, slides were placed into a humidified chamber and incubated overnight with a primary antibody diluted 1:300 in Dako Protein Block at 4°C. Antigen was detected using the SignalStain Boost immunohistochemistry detection reagent (Cat. No. 8114; Cell Signaling Technology, Beverly, MA), developed with diaminobenzidene (Dako), counterstained with hematoxylin (Sigma‐Aldrich), and mounted. Signal intensity was analyzed by Aperio ImageScope 12.1.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde, permeabilized with 1% Triton X‐100 for 15 minutes, then blocked with PBS containing 1% BSA for 1 hour. Cells were then incubated with primary antibody 1:300 in PBS containing 2.5 mM EDTA, 1% BSA, 0.1% Triton X‐100 overnight at 4°C. After washing with PBS, coverslips were incubated with Alexa Fluor‐conjugated secondary antibody (1:500) in 0.1 µg/mL 4´,6‐diamidino‐2‐phenylindole for 1 hour in the dark at room temperature. Coverslips were washed and mounted with FluorSave Reagent (Calbiochem, La Jolla, CA). Images were acquired using Keyence BZ‐800 microscope.
Reverse‐Transcription Polymerase Chain Reaction
RNA was extracted from livers using the RNeasy Mini Kit (Qiagen). cDNA was generated using the RNA reverse transcription (RT) kit (Cat. No. 4368814; Applied Biosystems). Quantitative real‐time RT‐polymerase chain reaction was performed in a CFX96 real‐time system (Bio‐Rad) using specific sense and antisense primers combined with iQ SYBR Green Supermix (Bio‐Rad) for 40 amplification cycles as follows: 5 seconds at 95 °C, 10 seconds at 57 °C, 30 seconds at 72 °C.
Cell Culture
Huh 7 cells( 23 ) (obtained from Dr. Charles Rice) were maintained in Dulbecco’s Modified Eagle’s Medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum, 50 U mL−1 penicillin, and 50 mg mL−1 streptomycin. Cells were transfected using Lipofectamine LTX transfection reagent (Invitrogen) according to the manufacturer’s protocol.
International Normalized Ratio/Prothrombin Time
Prothrombin time (PT) was measured using the Roche Diagnostics CoaguChek XS Professional Meter Kit.
Human Specimens
De‐identified human liver specimens from liver explants were obtained from the Liver Center Tissue Bank at the KUMC. All studies using human tissue samples were approved by the Human Subjects Committee of the KUMC.
Statistics
Results are expressed as mean ± SD. The Student t test, paired t test, Pearson's correlation, or one‐way analysis of variance with Bonferroni post hoc test was used for statistical analyses. P < 0.05 was considered significant.
Results
Single‐Cell RNA‐SEQ of Liver Cells Reveals Sex‐Specific Alcohol‐Induced Changes in Hepatocytes
To identify sex‐specific pathways affected by alcohol in individual subsets of liver cells, we performed single‐cell RNA (scRNA)‐Seq analysis of male and female mice fed a Lieber‐DeCarli control or alcohol liquid diet for 5 weeks (three mice/group). Different clusters in t‐distributed stochastic neighbor embedding and uniform manifold approximation and projection plots are shown in Fig. 1A. We identified two major hepatocyte clusters as well as clusters of nonparenchymal cell types (Fig. 1B). Cluster 1 (blue) showed higher expression of stearoyl‐coenzyme A desaturase 1 (Scd1) and other periportal hepatocyte markers (Fig. 1B), suggesting that this cluster represents zone I hepatocytes. We found that in alcohol‐fed mice there was an increase in the relative proportion of cluster 2 (Fig. 1B, red), in agreement with known data on the effect of alcohol on liver zonation.( 24 )
FIG. 1.
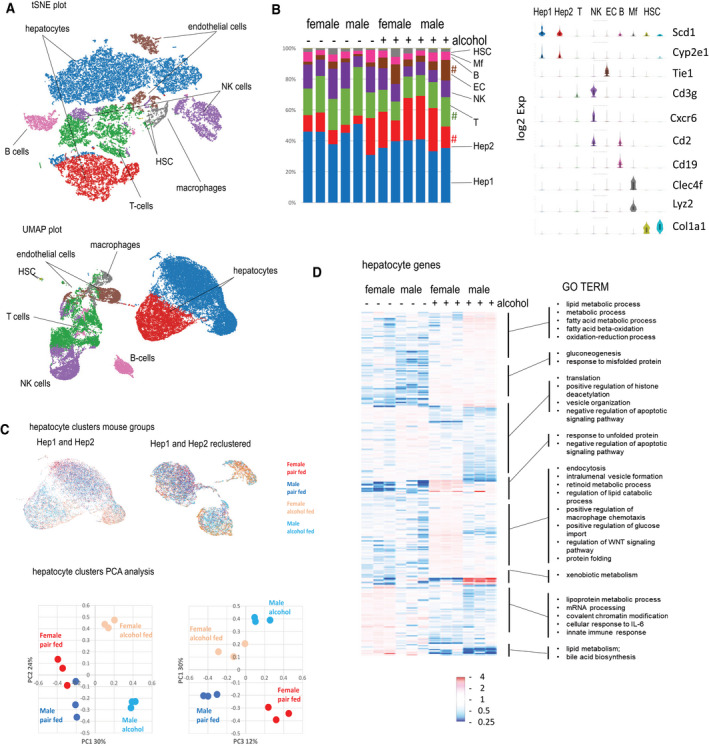
scRNA‐Seq identifies sex‐specific pathways induced by alcohol in the liver. (A‐F) Male and female mice were fed alcohol or control Lieber‐DeCarli liquid diet for 5 weeks; n = 3 mice per group. (A) tSNE (top) and UMAP (bottom) plots; (B) gene expression (log2) in main clusters and relative number of cells in each cluster across 12 samples; # P < 0.05 between pair‐fed and alcohol‐fed groups. (C) Hepatocyte clusters colored according to mouse groups. (C, left) Hep1 and Hep2 only; (C, right) Hep1 and Hep2 reclustered showing sex‐specific distribution. (C, bottom) PCA of hepatocyte (clusters 1 and 2) gene expression in 12 samples. (D) Top differentially regulated genes (P < 0.005) in hepatocyte clusters. GO term enrichment in differentially regulated genes in hepatocytes. Abbreviations: B, B‐cell; Cd, cluster of differentiation; Cle4f, C‐type lectin domain family 4 member F; Col1a1, collagen type 1 alpha 1 chain; Cxcr6, chemokine (C‐X‐C motif) receptor 6; EC, endothelial cell; HSC, hepatic stellate cell; IL, interleukin; Lyz, lysozyme; Mf, macrophage; NK, natural killer; PCA, principal component analysis; Scd1, Stearoyl‐CoA desaturase 1; Tie1, tyrosine kinase with immunoglobulin like and EGF like domains 1; tSNE, t‐distributed stochastic neighbor embedding; UMAP, uniform manifold approximation and projection plot.
Next, we examined the distribution of hepatocytes from four different groups (female or male pair or alcohol fed) in hepatocyte clusters before or after reclustering (Fig. 1C, top). We observed clear sex differences in the distribution. To confirm that this distribution was not dominated by a single animal, we performed principal component analysis on gene expression from hepatocytes in 12 individual mice (Fig. 1C). We found that male and female mice dramatically differ in their response to alcohol. We identified several subsets of genes differentially regulated in the two sexes (Fig. 1D). Genes that were up‐regulated in male but not female mice were involved in lipid and xenobiotic metabolism. On the other hand, genes up‐regulated in female but not male mice were involved in endocytosis, macrophage chemotaxis, and WNT signaling. Overall, we concluded that although alcohol can produce similar pathological changes in male and female livers (steatosis, inflammation, fibrosis, and loss of liver function( 1 , 7 , 25 , 26 , 27 )), the underlying mechanisms responsible for pathogenesis are different at the gene expression level (Fig. 1D).
Alcohol Promotes Hepatocyte Dedifferentiation in Male Mice
Our data suggest that in male mice, alcohol results in an increase in fatty acid synthesis, which is less pronounced in female mice (Fig. 2A). In males, we observed down‐regulation of genes associated with hepatocyte differentiation, such as Hnf4a, genes associated with liver synthetic function, including production of albumin (Alb) and coagulation factors (FVII and FVIII), and an increase in alpha‐fetoprotein (Afp), another marker of hepatocyte dedifferentiation. These changes were not present in female mice (Fig. 2B).
FIG. 2.
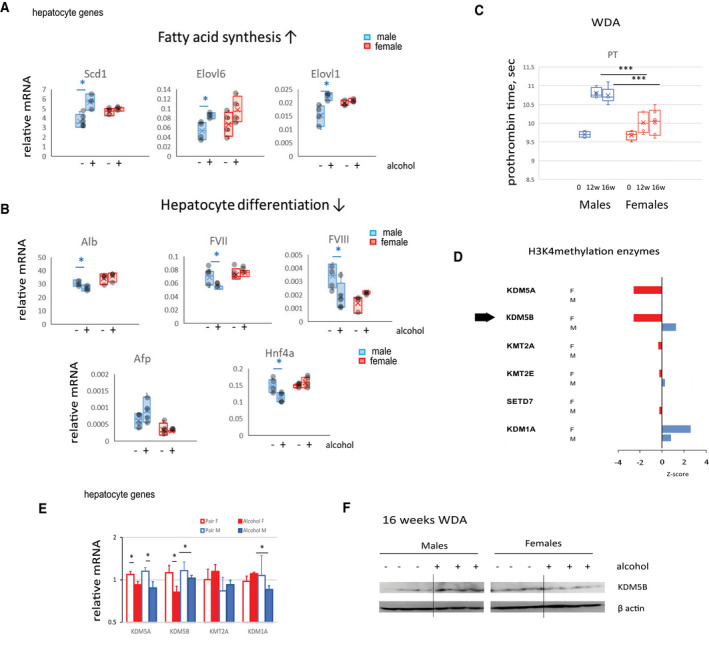
Alcohol induces male‐specific hepatocyte dedifferentiation and loss of synthetic liver function. (A,B) Relative gene expression in male and female mice fed alcohol or control Lieber‐DeCarli liquid diet for 3 weeks; n = 3‐4 mice per group; *P < 0.05. (C) PT in mice fed alcohol (WDA diet) for the indicated time; n = 4‐6 mice per group; ***P < 0.001. (D) Ingenuity pathway upstream regulator analysis. H3K4me enzymes predicted to regulate alcohol‐induced changes in male and female mice; P < 0.05. Z‐score indicates predicted activation (>2) or inhibition (≤2). (E) Gene expression in hepatocyte clusters of H3K4 epigenetic regulator expression in four groups of mice; data show mean ± SD; n = 3; *P < 0.05. (F) KDM5B protein expression in four groups of mice. Abbreviations: Afp, alpha‐fetoprotein; Elovl6, elongation of very long chain fatty acids protein 6; F, female; KMT, lysine methyltransferase; M, male; sec, seconds; SETD7, SET domain containing 7; w, weeks.
Reduced liver differentiation and liver synthetic function is evident in an increase of PT in alcohol‐fed mice (Fig. 2C). In agreement with gene expression data, we found that alcohol resulted in an increase of PT in both sexes but the effect was greater in male mice (Fig. 2C).
We analyzed upstream transcription regulators, transcription factors, and epigenetic regulators predicted to control alcohol‐induced changes in male and female mice by using Ingenuity Pathway Analysis (Qiagen). We found that several enzymes (methyltransferases and demethylases) specific for H3K4 methylation were predicted to regulate alcohol‐induced changes in hepatocytes, with P < 0.05. The predicted activation score or Z‐score is presented in Fig. 2D. Among these enzymes is the demethylase KDM5B, which showed significant overlap between its gene‐regulated network and alcohol‐induced differentially regulated genes in both female and male mice in hepatocytes. Hepatocyte KDM5B was predicted to be activated by alcohol in male mice and inhibited in female mice. Given its role in tumor development,( 9 , 10 ) KDM5B activation in male mice could contribute to alcohol‐induced hepatocyte dedifferentiation.
Moreover, we observed that hepatocyte messenger RNA (mRNA) levels of Kdm5b were reduced in female hepatocytes after alcohol exposure. We did not see a significant change in mRNA levels in male mice. At the protein level, we found an increase in KDM5B in male and a decrease in female mice after 16 weeks of alcohol feeding (Fig. 2E,F). This was in agreement with predicted male‐specific activation.
Kdm5b and Kdm5c Knockdown Results in Sex‐Specific Changes in Models of ALD
KDM5B and a related demethylase, KDM5C, have similar functions.( 28 , 29 ) They regulate a large variety of genes depending on context, i.e., cell type, stimulus state, or disease state.( 10 , 13 , 16 ) We observed that genes differentially regulated by alcohol in male and female mice overlapped with the KDM5B‐dependent gene set (Fig. 2D). Predicted activation scores in both sexes suggest that alcohol induces KDM5B and potentially KDM5C to regulate male‐specific changes induced by alcohol. Thus, knockdown of KDM5 demethylases might be expected to prevent alcohol‐induced changes in hepatocytes in male but not in female mice.
We therefore decided to evaluate the role of these two epigenetic regulators in sex‐dimorphic alcohol effects. We used AAV‐shRNA vectors to knockdown Kdm5b, Kdm5c, or both in the livers of mice fed an alcohol liquid diet for 3 weeks (Lieber‐DeCarli model; Fig. 3A) or fed alcohol for 16 weeks using a high‐fat Western diet in combination with alcohol in water (WDA model), as recently described by our group( 18 ) (Fig. 3B). Both diets have a similar amount of fat (36% and 42%, correspondingly) and result in comparable blood alcohol concentration.( 18 ) The WDA model, however, does not produce mortality like the Lieber‐DeCarli model, thus allowing for a longer duration of feeding (16 weeks with less than 1% mortality vs. 3 weeks with 10%‐20% mortality for the Lieber‐DeCarli model). Overall, the Lieber‐DeCarli model represents early stages of ALD while the WDA model represents later stages with more severe steatosis along with inflammation and fibrosis.
FIG. 3.
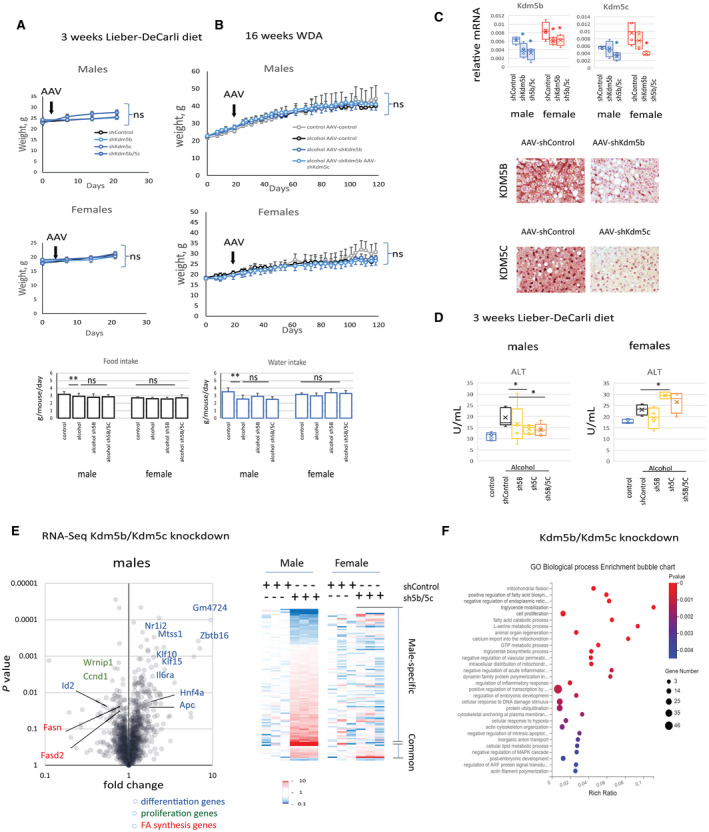
KDM5B and KDM5C role in alcohol‐fed mice is sex specific. (A) Male and female mice were fed the alcohol Lieber‐DeCarli liquid diet for 3 weeks; n = 5 mice per group. AAV as indicated was injected intraperitoneally at 1011 gc per mouse. (B) Male and female mice were fed ad libitum a Western diet and given either plain water (control) or water containing alcohol (alternating 10% and 20%) for 16 weeks; n = 4‐6 mice per group. AAV as indicated was injected intraperitoneally at 1011 gc per mouse. (C) KDM5B and KDM5C relative mRNA expression or protein expression by staining at ×20 magnification in livers of mice‐fed alcohol that received indicated AAV vectors; n = 4‐6 mice per group; *P < 0.05 compared to shControl group. (D) Serum ALT levels in mice fed the Lieber‐DeCarli liquid diet for 3 weeks; n = 5 mice per group; *P < 0.05. (E) Male and female mice were fed the alcohol Lieber‐DeCarli liquid diet for 3 weeks. Mice received AAV‐shControl at 2 × 1011 gc per mouse or AAV‐shKdm5b and AAV‐shKdm5c at 1 × 1011 gc of each virus per mouse; n = 3 mice per group. Differentially regulated genes in male mice. (F) GO term enrichment in differentially regulated genes from male mice. Data show mean ± SD. Abbreviations: gc, genome copies; ns, not significant.
Kdm5b and/or Kdm5c knockdown did not affect the weight gain or food and water intake in either protocol (Fig. 3A,B). AAV‐shRNA did successfully reduce levels of both mRNA and protein of KDM5B and KDM5C in the liver (Fig. 3C).
We found that Kdm5b/Kdm5c knockdown prevented alcohol‐induced ALT elevation in male mice but not in female mice (Fig. 3D). To find the targets of KDM5B/KDM5C demethylases in alcohol‐fed mice, we performed RNA‐Seq analysis (Fig. 3E). We observed that Kdm5b/Kdm5c knockdown resulted in gene expression changes of many genes in male mice. However, only a fraction of these genes were similarly regulated by Kdm5b/Kdm5c knockdown in female mice, suggesting a sex‐specific role of these factors in the liver (Fig. 3E, right panel). Gene Ontology (GO) analysis of differentially regulated genes revealed that Kdm5b/Kdm5c knockdown in male mice resulted in an increase in expression of genes associated with hepatocyte differentiation, e.g., Hnf4a, and a decrease in proliferation genes, such as cyclin D1 (Ccnd1) and inhibitor of DNA binding 2 (Id2; a negative regulator of hepatocyte differentiation), and a decrease in genes involved in fatty acid synthesis (Fig. 3E,F).
Alcohol Promotes Lipid Accumulation Through KDM5B and KDM5C in Male Mice
Because many genes affected by Kdm5b/Kdm5c knockdown were involved in lipid metabolism, we studied fat accumulation in alcohol‐fed mice. In the WDA model, alcohol resulted in massive fat accumulation in the livers of male mice. Kdm5b/Kdm5c knockdown prevented fat accumulation in male mice (Fig. 4A). In mice fed the Lieber‐DeCarli diet, we observed only a small amount of fat in the liver, but even this was reduced by Kdm5b/Kdm5c knockdown (Fig. 4B,C). In female mice, alcohol also increased histologic fat accumulation seen in hematoxylin and eosin (H&E) stains (Fig. 4A) and also produced a similar 2‐fold increase in liver triglyceride (TG) amount (Fig. 4C). However, overall levels of TG in female mice were significantly lower than in male mice (Fig. 4A,C), and Kdm5b/Kdm5c knockdown did not reduce lipid accumulation. This agreed with the observed decrease in the activity of the KDM5 demethylases in female mice.
FIG. 4.
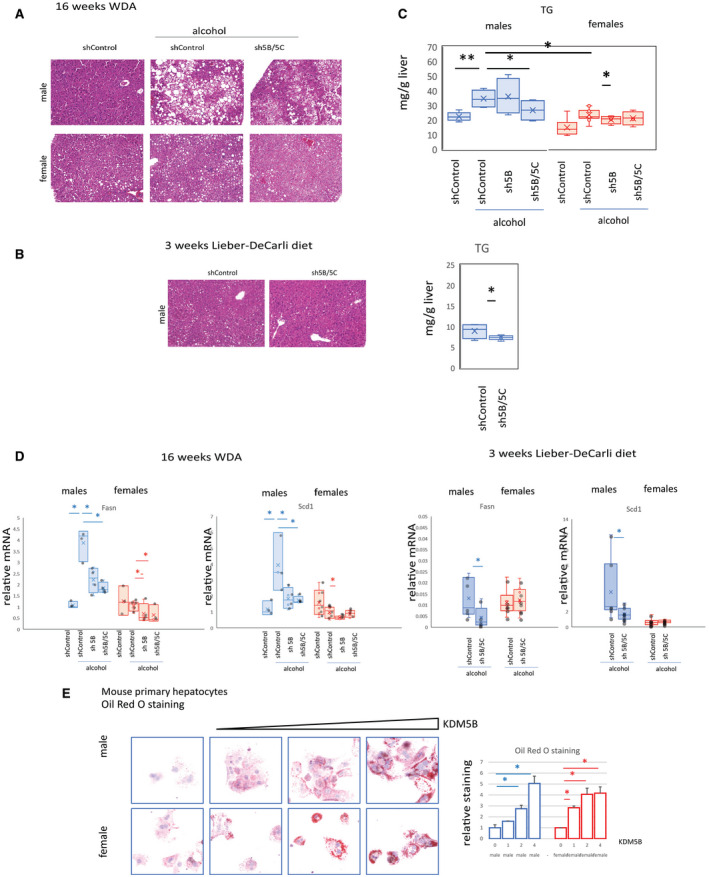
KDM5B and KDM5C regulate lipid metabolism in male mice. (A,B) H&E staining of liver section from mice as in Fig. 3 at ×10 magnification. (C) TG measurement; n = 4‐6 mice per group; *P < 0.05, **P < 0.01. (D) Relative mRNA expression; n = 4‐6 mice per group; *P < 0.05, **P < 0.01. (E) Oil Red O staining in mouse primary hepatocytes isolated from male or female mice overexpressing increasing amounts of KDM5B. (E, right) Relative staining intensity of Oil Red O staining in n = 3 independent experiments; *P < 0.05. Data show mean ± SD.
At the gene expression level, we found that the Kdm5b/Kdm5c knockdown effect on lipid accumulation was mediated by its effect on fatty acid synthesis, a mechanism specifically regulated by alcohol in male mice. In the WDA model, alcohol induced expression of fatty acid synthesis genes in male but not female mice in agreement with scRNA‐Seq data. Kdm5b/Kdm5c knockdown prevented this increase (Fig. 4D). Similar findings were observed in mice fed the Lieber‐DeCarli liquid diet (Fig. 4D).
We examined the effect of KDM5B overexpression on lipid accumulation in mouse primary hepatocytes isolated from male or female mice (Fig. 4E). We found that, as expected, KDM5B overexpression resulted in lipid accumulation in hepatocytes in a dose‐dependent manner and that this effect was sex independent. Taken together, the data suggest that alcohol promotes lipid accumulation in alcohol‐fed male mice through KDM5 up‐regulation.
Alcohol Suppresses Hnf4a Gene Expression Through KDM5B and KDM5C in Male But Not Female Mice
We found that alcohol suppresses Hnf4a gene expression specifically in male mice (Fig. 2). Kdm5b/Kdm5c knockdown reproduced this sex‐specific effect and resulted in up‐regulation of Hnf4a in male but not female mice (Fig. 3E). Because HNF4α is a master regulator of hepatocyte differentiation, we aimed to investigate the mechanism of KDM5B/KDM5C‐dependent regulation of Hnf4a gene expression in the presence of alcohol.
In mice fed the Lieber‐DeCarli alcohol diet, we found that either Kdm5b or Kdm5c knockdown (Fig. 5A, left) or the knockdown of both enzymes together (Fig. 5A, right) increased Hnf4a expression specifically in male mice. In the WDA model, we found that alcohol decreased Hnf4a gene expression in male mice more than in female mice and that Kdm5b/Kdm5c knockdown prevented this decrease (Fig. 5B), suggesting that this mechanism is independent of the ALD model.
FIG. 5.
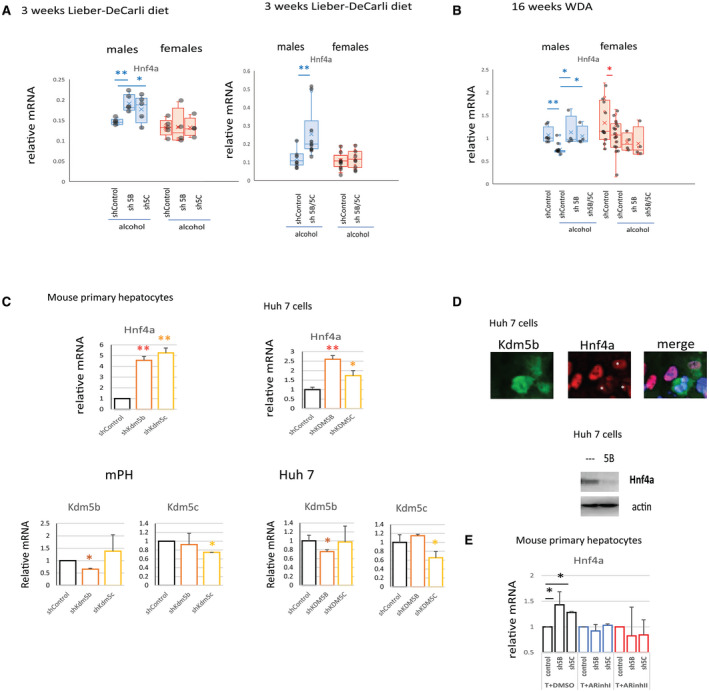
KDM5B and KDM5C suppress Hnf4a expression in alcohol‐fed male but not female mice. (A) Relative mRNA expression in livers of mice as in Fig. 3A; n > 5 mice per group; *P < 0.05, **P < 0.01. (B) Relative mRNA expression in livers of mice as in Fig. 3B; n = 4‐6 mice per group; *P < 0.05, **P < 0.01. (C) Relative mRNA expression in mouse primary hepatocytes or Huh 7 cells that received shControl vector or vector specific for Kdm5b or Kdm5c as indicated; n = 4 independent experiments; *P < 0.05, **P < 0.01. (D) HNF4α protein levels by western blot analysis or by immunofluorescence staining at 40× magnification in Huh 7 cells overexpressing KDM5B. (E) Mouse primary hepatocytes expressing control shRNA (control) or shRNA specific for Kdm5b (sh5B) or Kdm5c (sh5C) were treated with two different AR inhibitors. The AR DBD inhibitor, VPC‐14449, blocks the DNA‐binding domain of AR (ARinhI). A second inhibitor, ailanthone, disrupts the interaction between AR and the chaperones HSP90, HSP70, and HSP40, which leads to AR ubiquitination and degradation (ARinhII). Relative mRNA expression in n = 4 independent experiments; *P < 0.05. Data show mean ± SD. Abbreviations: ARinh, androgen receptor inhibitor; DBD, DNA‐binding domain; DMSO, dimethyl sulfoxide; mPH, mouse primary hepatocytes; T, testosterone.
Next, we confirmed that the effect of Kdm5b/Kdm5c knockdown in the whole liver was mediated by its effect in hepatocytes. We studied the effect of Kdm5b/Kdm5c knockdown on Hnf4a up‐regulation in vitro in mouse primary hepatocytes and Huh 7 cells (Fig. 5C). We found that either Kdm5b or Kdm5c knockdown increased Hnf4a expression. In contrast, KDM5B overexpression resulted in HNF4α down‐regulation (Fig. 5D). We found that cells overexpressing KDM5B showed a dramatic reduction of HNF4α protein level (Fig. 5D).
KDM5B is known to bind AR and regulate AR‐dependent gene expression.( 30 ) To test the role of AR in male‐specific KDM5B/5C‐dependent regulation, we tested the effect of two AR inhibitors in vitro in mouse primary hepatocytes (Fig. 5E). We found that AR inhibition in the presence of testosterone could prevent KDM5B/5C‐dependent suppression of Hnf4a gene expression, suggesting that AR signaling is involved in male‐specific KDM5B and KDM5C activation and Hnf4a down‐regulation.
Alcohol Alters KDM5B and KDM5C Promoter Binding in a Sex‐Specific Way
KDM5B and KDM5C are histone demethylases that can suppress gene expression through promoter binding and histone demethylation. Next, we tested whether KDM5B and KDM5C bind Hnf4a promoter. We found that both KDM5B and KDM5C were recruited to Hnf4a promoter after alcohol exposure (16 weeks WDA) in male but not female mice (Fig. 6A). This recruitment resulted in a decrease of histone H3K4me3 methylation, which correlated with reduced gene expression specifically in male mice (Fig. 6A). Similarly, we observed an increase in KDM5B binding to Hnf4a promoter in male but not female mice fed the Lieber‐DeCarli diet compared to pair‐fed controls. Kdm5b/Kdm5c knockdown resulted in decreased binding of KDM5B to Hnf4a promoter (Fig. 6B), suggesting that KDM5B and KDM5C control Hnf4a expression in the presence of alcohol through increased binding to its promoter specifically in male mice.
FIG. 6.
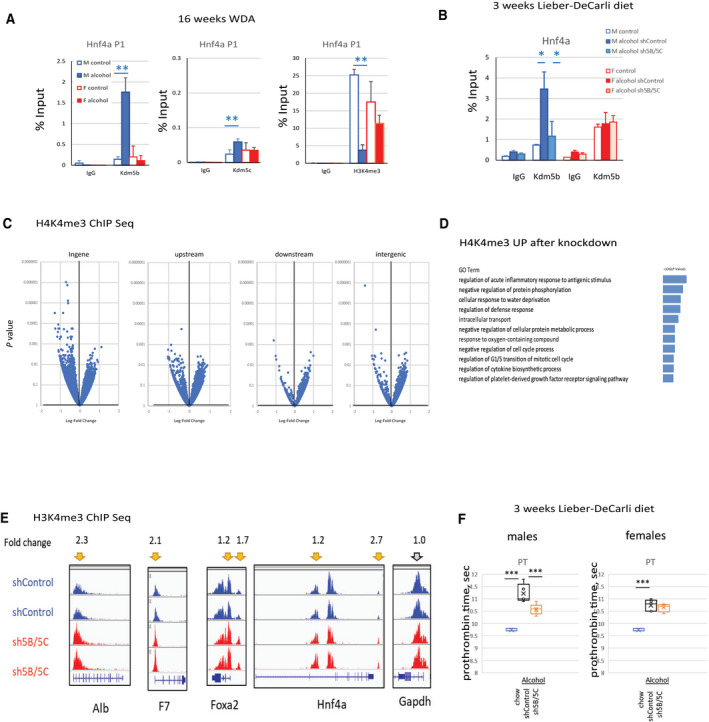
Alcohol‐induced KDM5B and KDM5C altered promoter binding to suppress Hnf4a expression and hepatocyte differentiation in male mice. (A) ChIP using antibodies specific to KDM5B, KDM5C, H3K4me3, or IgG as a negative control in livers from male and female mice fed control or alcohol diet; n = 4‐6 mice per group; **P < 0.01. (B) KDM5B ChIP in livers of mice as in Fig. 3A; n = 5 mice per group; *P < 0.05. (C) ChIP‐Seq analysis of whole livers from alcohol‐fed male mice as in Fig. 3B. Mice received AAV‐shControl at 2 × 1011 gc per mouse or AAV‐shKdm5b and AAV‐shKdm5c at 1 × 1011 gc of each virus per mouse; n = 3 mice per group. Volcano plot of differentially regulated regions. (D) GO term enrichment in genes associated with regions that showed increased H3K4 methylation after Kdm5b and Kdm5c knockdown. (E) Examples of differentially regulated regions. (F) PT in mice fed the Lieber‐DeCarli liquid diet as in Fig. 3A and chow control mice; n = 4‐6 mice per group. ***P < 0.001. Data show mean ± SD. Abbreviations: F, female, Foxa2, forkhead box A2; Gapdh, glyceraldehyde 3‐phosphate dehydrogenase; gc, genome copies; IgG, immunoglobulin; M, male; sec, seconds.
To further determine the targets of KDM5B/KDM5C in alcohol‐fed mouse livers, we studied global epigenetic changes induced by Kdm5b/Kdm5c knockdown in the liver after alcohol exposure by using ChIP‐Seq analysis of H3K4me3 modifications (Fig. 6C). We observed multiple changes in H3K4me3 peaks. Increased promoter histone methylation was observed in genes related to regulation of inflammatory response, cell‐cycle processes, metabolism, coagulation, and others (Fig. 6D).
These data suggest that KDM5B and KDM5C directly bind and demethylate promoters associated with hepatocyte differentiation and liver synthetic function. We observed an increase of histone H3K4 methylation after Kdm5b/Kdm5c knockdown at promoters of genes, such as Alb, coagulation factor F7, and the transcription factors involved in hepatocyte differentiation, forkhead box A2 (Foxa2) and Hnf4a (Fig. 6E).
In addition, as predicted, we observed that Kdm5b/Kdm5c knockdown resulted in reduced PT in male but not female mice (Fig. 6F), suggesting that alcohol‐mediated loss of liver synthetic function in male mice is KDM5B/KDM5C dependent.
KDM5 Demethylase Inhibition Recapitulates the Effect of Kdm5b and Kdm5c Knockdown
Several KDM5 inhibitors were developed with different specificity and affinity.( 12 , 31 ) We tested four inhibitors in vitro in mouse primary hepatocytes and assessed their effects on gene expression of Hnf4a, fatty acid synthase (Fasn), and peroxisome proliferator‐activated receptor alpha (Ppara), a gene involved in fatty acid β‐oxidation.
We found that after a 24‐hour treatment of freshly isolated hepatocytes, KDM5A‐in‐1 (5in1) and to a lesser extent Kdm5‐C70 (C70) were effective in inducing Hnf4a and Ppara gene expression and suppressing Fasn expression. 2‐4(4‐methylphenyl)‐1,2‐benzisothiazol‐3(2H)‐one (PBIT) was effective in inducing Hnf4a gene expression and suppressing Fasn expression. These three inhibitors were specific to KDM5 demethylases, with highest affinity for KDM5A, KDM5B, and KDM5C.( 12 , 31 , 32 ) The inhibitor AS8351 (AS), an alpha‐ketoglutaric acid (α‐KG) competitor not specific to KDM5 demethylases, did not show significant gene expression changes (Fig. 7A).
FIG. 7.
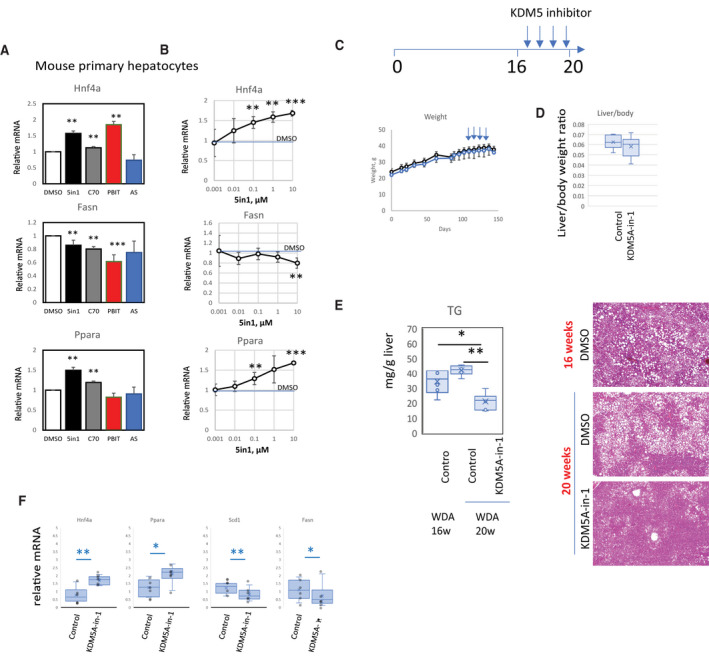
Pan‐KDM5 demethylase inhibition recapitulates the effect of Kdm5b and Kdm5c knockdown. (A) Mouse primary hepatocytes were treated with 10 µM of indicated inhibitors (5in1, C70, PBIT, AS) or DMSO control for 24 hours. Relative mRNA levels of Hnf4a, Fasn, and Ppara; n = 4 independent experiments; **P < 0.01, ***P < 0.001. (B) KDM5A‐in‐1 inhibitor was added at indicated concentrations to mouse primary hepatocytes for 24 hours. Relative mRNA levels of Hnf4a, Fasn, and Ppara; n = 4 independent experiments; **P < 0.01, ***P < 0.001. (C) Mice were fed a WDA diet for 16 weeks and then given weekly injections of DMSO/PBS or 5 mg/kg of KDM5A‐in‐1. (D) Liver/body weight ratios at 20 weeks. (E) TG measurement and H&E staining at ×10 magnification of male mice at 16 weeks and 20 weeks. (F) Relative mRNA expression in these mice at 20 weeks; n = 6 mice per group; *P < 0.05, **P < 0.01. Data show mean ± SD. Abbreviations: 5in1, KDM5A‐in‐1; AS, AS8351; C70, KDM5‐C70; DMSO, dimethyl sulfoxide; PBIT, 2‐4(4‐methylphenyl)‐1,2‐benzisothiazol‐3(2H)‐one (PBIT); w, weeks.
In addition, KDM5A‐in‐1 showed a dose response, with 100 nM, 1 µM, and 10 µM doses all showing significant elevations of Hnf4a and Ppara gene expression (Fig. 7B).
Next, we tested KDM5A‐in‐1 inhibitor in vivo in male mice. We treated mice with four weekly intraperitoneal 5 mg/kg injections starting after 16 weeks of alcohol feeding (Fig. 7C). Mice did not show any difference in weight gain. However, we observed that inhibitor treatment was efficiently suppressing lipid accumulation compared to vehicle control at 20 weeks (Fig. 7D,E). In addition, treated mice showed a significantly lower amount of liver fat compared to the 16‐week group, suggesting that inhibition of KDM5 demethylases can reverse lipid accumulation in alcohol‐fed mice.
We observed that inhibitor treatment was efficient in inducing Hnf4a and Ppara gene expression and suppressing Scd1 and Fasn expression in vivo (Fig. 7F), recapitulating the effect of Kdm5b and Kdm5c knockdown.
KDM5B is Involved in HCC Development in Males
Hepatocellular carcinoma (HCC) is a male‐predominant disease,( 33 , 34 , 35 ) often associated with a loss of hepatocyte differentiation factors, including HNF4α.( 36 , 37 , 38 ) We thus hypothesized that KDM5B‐dependent HNF4α regulation can be involved in HCC development in male mice. To test this hypothesis, we knocked down Kdm5b in 6‐month‐old male mice that received a single diethylnitrosamine injection at 2 weeks of age (Fig. 8A). We observed that Kdm5b knockdown reduced tumor development at 8 months in these mice (Fig. 8A,B). We observed that KDM5B protein levels were higher in tumors compared to adjacent tissue (Fig. 8C). Reduced KDM5B levels in Kdm5b knockdown mice correlated with an increase in HNF4α protein and mRNA expression (Fig. 8C,D) and reduced tumor proliferation (Fig. 8D).
FIG. 8.
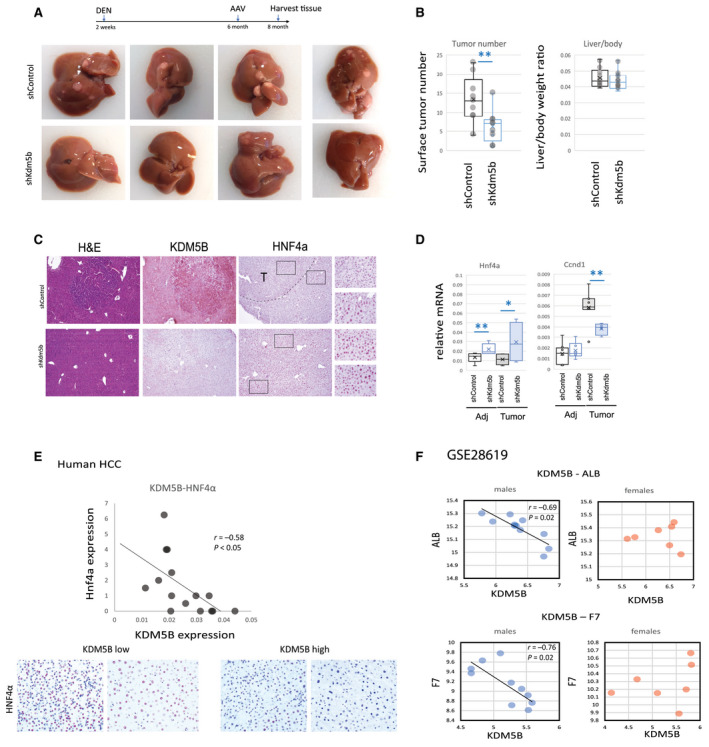
KDM5B promotes HNF4α loss and development of HCC. (A‐D) Mice received a single injection of DEN (10 mg/kg) at 2 weeks of age. At 6 months of age, mice were treated with AAV‐shControl or AAV‐shKdm5b at 2 × 1011 gc per mouse. (B) Tumor number and liver/body weight ratio in these mice, n = 9 mice per group; **P < 0.01. (C) Representative images of H&E staining or staining for KDM5B and HNF4α proteins at ×10 magnification. (D) Relative mRNA levels in tumor and adjacent tissues of these mice; n = 9 mice per group (liver tissue), n = 3‐6 mice (tumor tissue); *P < 0.05, **P < 0.01. (E) Human transplant explants HCC sections were analyzed for HNf4α and KDM5B expression; n = 16; images are at ×20 magnification. (F) Gene expression analysis of GSE28619. Male and female samples were analyzed separately. Data show mean ± SD. Abbreviations: Adj, adjacent; DEN, diethylnitrosamine; gc, genome copies.
Next, we confirmed our findings in human liver samples. We used human liver transplant explants from the KUMC Liver Bank to analyze gene and protein expression.
We found that KDM5B expression negatively correlated with HNF4α expression in male HCC samples (Fig. 8E). Samples with high KDM5B expression showed low HNF4α staining, while samples with low KDM5B expression showed high HNF4α staining (Fig. 8E).
Taken together, these data suggest that KDM5B is activated in male HCC in a similar way as in the presence of alcohol and contributes to male HCC development through regulation of hepatocyte differentiation and proliferation.
Finally, we confirmed our observations in liver samples from patients with AH (GSE28619( 39 )). We found strong negative correlation between KDM5B expression and expression of ALB and F7 in male samples. In contrast, no correlation was present in female samples (Fig. 8F). These data suggest that KDM5 demethylases are involved in male‐specific dedifferentiation and loss of liver function in human disease as well.
Discussion
ALD is a major cause of alcohol‐related mortality.( 1 , 40 , 41 , 42 , 43 ) Chronic alcohol abuse underlies the pathogenesis of a spectrum of diseases, ranging from steatosis to more severe forms of liver injury, including AH, fibrosis, cirrhosis, and HCC. The specific mechanisms responsible for ALD development and progression are not fully understood.
In the past, sex differences in ALD have been mainly attributed to differences in alcohol metabolism.( 5 , 6 , 7 ) However, recent studies revealed that in both mice and humans males and females differ not only in lipid‐related pathways but also in innate and adaptive immunity, fibrosis signaling, growth factor receptor signaling, and cancer pathways.( 44 ) Gonadectomy and ovariectomy experiments suggest that these pathways are in part regulated by sex hormone signaling, suggesting that ALD development might be affected by similar mechanisms.
In this study, we found that liver molecular pathways are altered by alcohol consumption differently in male and female mice. We identified several differentially regulated pathways in hepatocytes and were able to associate these changes with corresponding upstream regulators. We found that H3K4‐specific demethylase enzymes, particularly the demethylase KDM5B, are among the top regulators of differentially regulated pathways in male and female mice. We found that in alcohol‐fed mice, KDM5B and another demethylase, KDM5C, have sex‐specific roles in the liver after alcohol exposure. Specifically in male mice, KDM5B and KDM5C are involved in alcohol‐induced dedifferentiation and Hnf4a down‐regulation and an increase in fatty acid synthesis. This mechanism is not present in female mice.
While we were able to show that KDM5B and KDM5C are key regulators of the set of sex differential alcohol‐regulated genes, we did not detect sex‐specific differences in either the amount or enzymatic activity of these epigenetic regulator proteins. It is thus not clear why these demethylases are activated differently in male and female mice.
We used two mouse models of ALD, short duration Lieber‐DeCarli liquid diet feeding (36% calories from fat) and 16 weeks of high‐fat diet feeding (43% calories from fat) in combination with alcohol in drinking water.( 18 ) Both diets have similar amounts of fat, no added fructose or high cholesterol, and result in comparable blood alcohol concentration, but the 16‐week WDA model produces more severe steatosis along with inflammation and fibrosis. The models are complementary in that they represent either mild alcohol‐associated steatosis or more severe alcohol‐associated steatohepatitis. We found similar effects of Kdm5b/Kdm5c knockdown in these two models. We observed the male‐specific effect on Hnf4a gene expression and fatty acid synthesis genes in both models. We observed small differences between the models in expression of fatty acid metabolism genes, possibly due to differences in fat composition in these two models and/or the duration of the feeding.
Our data suggest that the effect we observed in male mice can be largely attributed to Kdm5b/Kdm5c knockdown resulting in changes in hepatocyte Hnf4a expression and hepatocyte differentiation. HNF4α is known to promote hepatocyte differentiation, reduce lipid accumulation and proliferation, and protect from cancer development.( 45 , 46 ) Thus, it is likely that the effect of Kdm5b/Kdm5c knockdown is in part due to Hnf4a up‐regulation. However, several other genes induced by Kdm5b/Kdm5c knockdown are known to be HNF4α independent, including coagulation factors. These targets are likely controlled by Kdm5b/Kdm5c directly through histone methylation.
We confirmed that the relationship between KDM5B and hepatocyte differentiation is male specific in human liver samples from patients with AH and HCC, suggesting that this mechanism is relevant in human disease.
Moreover, we established that KDM5B regulates male HCC development in a mouse model. In an HCC model, we observed that KDM5B expression in tumors was up‐regulated similarly to alcohol condition. We observed a strong negative correlation between KDM5B and HNF4α expression in both cases. Taken together, targeting these enzymes in males can be a promising strategy for preventing alcohol‐induced loss of HNF4α and potentially HCC development. These data are in agreement with a reported role of KDM5 demethylases in HCC, a male‐predominant disease.( 11 , 12 )
It is still unclear why alcohol promotes KDM5B and KDM5C activation in male but not female hepatocytes. One of the possible mechanisms is hormone receptor signaling. Recent studies reported that sex bias in liver gene expression (including Hnf4a expression) is mediated by epigenetic mechanisms.( 6 , 7 , 47 , 48 ) Global ablation of AR or estrogen receptor 1 alter sex‐biased epigenetic marks, suggesting that sex hormone signaling is an upstream regulator of male‐biased and female‐biased gene expression.( 47 ) We observed that AR inhibition ablated the effect of Kdm5b and Kdm5c knockdown on Hnf4a expression, suggesting that AR might be involved in KDM5 demethylase activation in male mice.
Overall, our data suggest that lysine demethylases KDM5B and KDM5C contribute to sex differences in ALD progression. The key message is that different therapeutic approaches may be required in male and female patients to best prevent and treat ALD. Future studies to identify the mechanisms by which these demethylases are linked to alcohol will be necessary in order to optimize these approaches.
Supported by the National Institute on Alcoholism and Alcohol Abuse (grants AA027586 to I.T. and AA012863 to S.A.W.) and Veteran Affairs Merit Award (I01BX004694 to S.A.W.)
Potential conflict of interest: Nothing to report.
References
- 1. Pang JXQ, Ross E, Borman MA, Zimmer S, Kaplan GG, Heitman SJ, et al. Risk factors for mortality in patients with alcoholic hepatitis and assessment of prognostic models: a population‐based study. Can J Gastroenterol Hepatol 2015;29:131‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang GQ, Karatayev O, Boorgu D, Leibowitz SF. CCL2/CCR2 chemokine system in embryonic hypothalamus: involvement in sexually dimorphic stimulatory effects of prenatal ethanol exposure on peptide‐expressing neurons. Neuroscience 2020;424:155‐171. Erratum in: Neuroscience 2020;442:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eagon PK, Willett JE, Seguiti SM, Appler ML, Gavaler JS, Van Thiel DH. Androgen‐responsive functions of male rat liver. Effect of chronic alcohol ingestion. Gastroenterology 1987;93:1162‐1169. [DOI] [PubMed] [Google Scholar]
- 4. Penaloza CG, Cruz M, Germain G, Jabeen S, Javdan M, Lockshin RA, et al. Higher sensitivity of female cells to ethanol: methylation of DNA lowers Cyp2e1, generating more ROS. Cell Commun Signal 2020;18:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Simon FR, Fortune J, Iwahashi M, Sutherland E. Sexual dimorphic expression of ADH in rat liver: importance of the hypothalamic‐pituitary‐liver axis. Am J Physiol Gastrointest Liver Physiol 2002;283:G646‐G655. [DOI] [PubMed] [Google Scholar]
- 6. Muller MF, Kendall TJ, Adams DJ, Zhou Y, Arends MJ. The murine hepatic sequelae of long‐term ethanol consumption are sex‐specific and exacerbated by Aldh1b1 loss. Exp Mol Pathol 2018;105:63‐70. [DOI] [PubMed] [Google Scholar]
- 7. Li S‐Q, Wang P, Wang D‐M, Lu H‐J, Li R‐F, Duan L‐X, et al. Molecular mechanism for the influence of gender dimorphism on alcoholic liver injury in mice. Hum Exp Toxicol 2019;38:65‐81. [DOI] [PubMed] [Google Scholar]
- 8. Wynder C, Stalker L, Doughty ML. Role of H3K4 demethylases in complex neurodevelopmental diseases. Epigenomics 2010;2:407‐418. [DOI] [PubMed] [Google Scholar]
- 9. Fu YD, Huang MJ, Guo JW, You YZ, Liu HM, Huang LH, et al. Targeting histone demethylase KDM5B for cancer treatment. Eur J Med Chem 2020;208:112760. [DOI] [PubMed] [Google Scholar]
- 10. Han M, Xu W, Cheng P, Jin H, Wang X. Histone demethylase lysine demethylase 5B in development and cancer. Oncotarget 2017;8:8980‐8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jose A, Shenoy GG, Sunil Rodrigues G, Kumar NAN, Munisamy M, Thomas L, et al. Histone demethylase KDM5B as a therapeutic target for cancer therapy. Cancers (Basel) 2020;12:2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang J, Labadie S, Zhang B, Ortwine DF, Patel S, Vinogradova M, et al. From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors. Bioorg Med Chem Lett 2017;27:2974‐2981. [DOI] [PubMed] [Google Scholar]
- 13. Mocavini I, Pippa S, Licursi V, Paci P, Trisciuoglio D, Mannironi C, et al. JARID1B expression and its function in DNA damage repair are tightly regulated by miRNAs in breast cancer. Cancer Sci 2019;110:1232‐1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen K, Luan X, Liu Q, Wang J, Chang X, Snijders AM, et al. Drosophila histone demethylase KDM5 regulates social behavior through immune control and gut microbiota maintenance. Cell Host Microbe 2019;25:537‐552.e538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaillard S, Charasson V, Ribeyre C, Salifou K, Pillaire M‐J, Hoffmann J‐S, et al. KDM5A and KDM5B histone‐demethylases contribute to HU‐induced replication stress response and tolerance. Biol Open 2021;10:bio057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu B, Kumar R, Chao HP, Mehmood R, Ji Y, Tracz A, et al. Evidence for context‐dependent functions of KDM5B in prostate development and prostate cancer. Oncotarget 2020;11:4243‐4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo F, Zheng K, Benede‐Ubieto R, Cubero FJ, Nevzorova YA. The Lieber‐Decarli diet‐a flagship model for experimental alcoholic liver disease. Alcohol Clin Exp Res 2018;42:1828‐1840. [DOI] [PubMed] [Google Scholar]
- 18. Schonfeld M, O'Neil M, Villar MT, Artigues A, Averilla J, Gunewardena S, et al. A Western diet with alcohol in drinking water model recapitulates features of alcohol‐associated liver disease in mice. Alcohol Clin Exp Res 2021;45:1980‐1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Troutman TD, Bennett H, Sakai M, Seidman JS, Heinz S, Glass CK. Purification of mouse hepatic non‐parenchymal cells or nuclei for use in ChIP‐seq and other next‐generation sequencing approaches. STAR Protoc 2021;2:100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Z, Zhao J, Tikhanovich I, Kuravi S, Helzberg J, Dorko K, et al. Serine 574 phosphorylation alters transcriptional programming of FOXO3 by selectively enhancing apoptotic gene expression. Cell Death Differ 2016;23:583‐595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tikhanovich I, Zhao J, Olson J, Adams A, Taylor R, Bridges B, et al. Protein arginine methyltransferase 1 modulates innate immune responses through regulation of peroxisome proliferator‐activated receptor gamma‐dependent macrophage differentiation. J Biol Chem 2017;292:6882‐6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao J, Adams A, Roberts B, O'Neil M, Vittal A, Schmitt T, et al. Protein arginine methyl transferase 1‐ and Jumonji C domain‐containing protein 6‐dependent arginine methylation regulate hepatocyte nuclear factor 4 alpha expression and hepatocyte proliferation in mice. Hepatology 2018;67:1109‐1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones CT, Catanese MT, Law LMJ, Khetani SR, Syder AJ, Ploss A, et al. Real‐time imaging of hepatitis C virus infection using a fluorescent cell‐based reporter system. Nat Biotechnol 2010;28:167‐171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu S, Yeh TH, Singh VP, Shiva S, Krauland L, Li H, et al. beta‐catenin is essential for ethanol metabolism and protection against alcohol‐mediated liver steatosis in mice. Hepatology 2012;55:931‐940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghosh Dastidar S, Warner JB, Warner DR, McClain CJ, Kirpich IA. Rodent models of alcoholic liver disease: role of binge ethanol administration. Biomolecules 2018;8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eagon PK. Alcoholic liver injury: influence of gender and hormones. World J Gastroenterol 2010;16:1377‐1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Belknap JK, Crabbe JC, Young ER. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology 1993;112:503‐510. [DOI] [PubMed] [Google Scholar]
- 28. Glanzner WG, Gutierrez K, Rissi VB, de Macedo MP, Lopez R, Currin L, et al. Histone lysine demethylases KDM5B and KDM5C modulate genome activation and stability in porcine embryos. Front Cell Dev Biol 2020;8:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shao G‐B, Chen J‐C, Zhang L‐P, Huang P, Lu H‐Y, Jin J, et al. Dynamic patterns of histone H3 lysine 4 methyltransferases and demethylases during mouse preimplantation development. In Vitro Cell Dev Biol Anim 2014;50:603‐613. [DOI] [PubMed] [Google Scholar]
- 30. Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, et al. JARID1B is a histone H3 lysine 4 demethylase up‐regulated in prostate cancer. Proc Natl Acad Sci U S A 2007;104:19226‐19231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pippa S, Mannironi C, Licursi V, Bombardi L, Colotti G, Cundari E, et al. Small molecule inhibitors of KDM5 histone demethylases increase the radiosensitivity of breast cancer cells overexpressing JARID1B. Molecules 2019;24:1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Horton J, Liu XU, Gale M, Wu L, Shanks J, Zhang X, et al. Structural Basis for KDM5A histone lysine demethylase inhibition by diverse compounds. Cell Chem Biol 2016;23:769‐781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Joshi K, Kohli A, Manch R, Gish R. Alcoholic liver disease: high risk or low risk for developing hepatocellular carcinoma? Clin Liver Dis 2016;20:563‐580. [DOI] [PubMed] [Google Scholar]
- 34. Sanyal AJ, Yoon SK, Lencioni R. The etiology of hepatocellular carcinoma and consequences for treatment. Oncologist 2010;15(Suppl. 4):14‐22. [DOI] [PubMed] [Google Scholar]
- 35. Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis 2007;27:55‐76. [DOI] [PubMed] [Google Scholar]
- 36. Désert R, Rohart F, Canal F, Sicard M, Desille M, Renaud S, et al. Human hepatocellular carcinomas with a periportal phenotype have the lowest potential for early recurrence after curative resection. Hepatology 2017;66:1502‐1518. [DOI] [PubMed] [Google Scholar]
- 37. Ramesh V, Ganesan K. Integrative functional genomic delineation of the cascades of transcriptional changes involved in hepatocellular carcinoma progression. Int J Cancer 2016;139:1586‐1597. [DOI] [PubMed] [Google Scholar]
- 38. Walesky C, Edwards G, Borude P, Gunewardena S, O'Neil M, Yoo B, et al. Hepatocyte nuclear factor 4 alpha deletion promotes diethylnitrosamine‐induced hepatocellular carcinoma in rodents. Hepatology 2013;57:2480‐2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Affò S, Dominguez M, Lozano JJ, Sancho‐Bru P, Rodrigo‐Torres D, Morales‐Ibanez O, et al. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut 2013;62:452‐460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anstee QM, Daly AK, Day CP. Genetics of alcoholic liver disease. Semin Liver Dis 2015;35:361‐374. [DOI] [PubMed] [Google Scholar]
- 41. Osna NA, Donohue TM Jr, Kharbanda KK. Alcoholic liver disease: pathogenesis and current management. Alcohol Res 2017;38:147‐161. [PMC free article] [PubMed] [Google Scholar]
- 42. Stickel F, Hampe J. Genetic determinants of alcoholic liver disease. Gut 2012;61:150‐159. [DOI] [PubMed] [Google Scholar]
- 43. Kamath PS, Kim WR; Advanced Liver Disease Study Group . The model for end‐stage liver disease (MELD). Hepatology 2007;45:797‐805. [DOI] [PubMed] [Google Scholar]
- 44. Kurt Z, Barrere‐Cain R, LaGuardia J, Mehrabian M, Pan C, Hui ST, et al. Tissue‐specific pathways and networks underlying sexual dimorphism in non‐alcoholic fatty liver disease. Biol Sex Differ 2018;9:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Walesky C, Apte U. Role of hepatocyte nuclear factor 4alpha (HNF4alpha) in cell proliferation and cancer. Gene Expr 2015;16:101‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walesky C, Gunewardena S, Terwilliger EF, Edwards G, Borude P, Apte U. Hepatocyte‐specific deletion of hepatocyte nuclear factor‐4alpha in adult mice results in increased hepatocyte proliferation. Am J Physiol Gastrointest Liver Physiol 2013;304:G26‐G37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. AlOgayil N, Bauermeister K, Galvez JH, Venkatesh VS, Zhuang QK, Chang ML, et al. Distinct roles of androgen receptor, estrogen receptor alpha, and BCL6 in the establishment of sex‐biased DNA methylation in mouse liver. Sci Rep 2021;11:13766. Erratum in: Sci Rep 2021;11:22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lau‐Corona D, Bae WK, Hennighausen L, Waxman DJ. Sex‐biased genetic programs in liver metabolism and liver fibrosis are controlled by EZH1 and EZH2. PLoS Genet 2020;16:e1008796. [DOI] [PMC free article] [PubMed] [Google Scholar]


