Fig. 12.
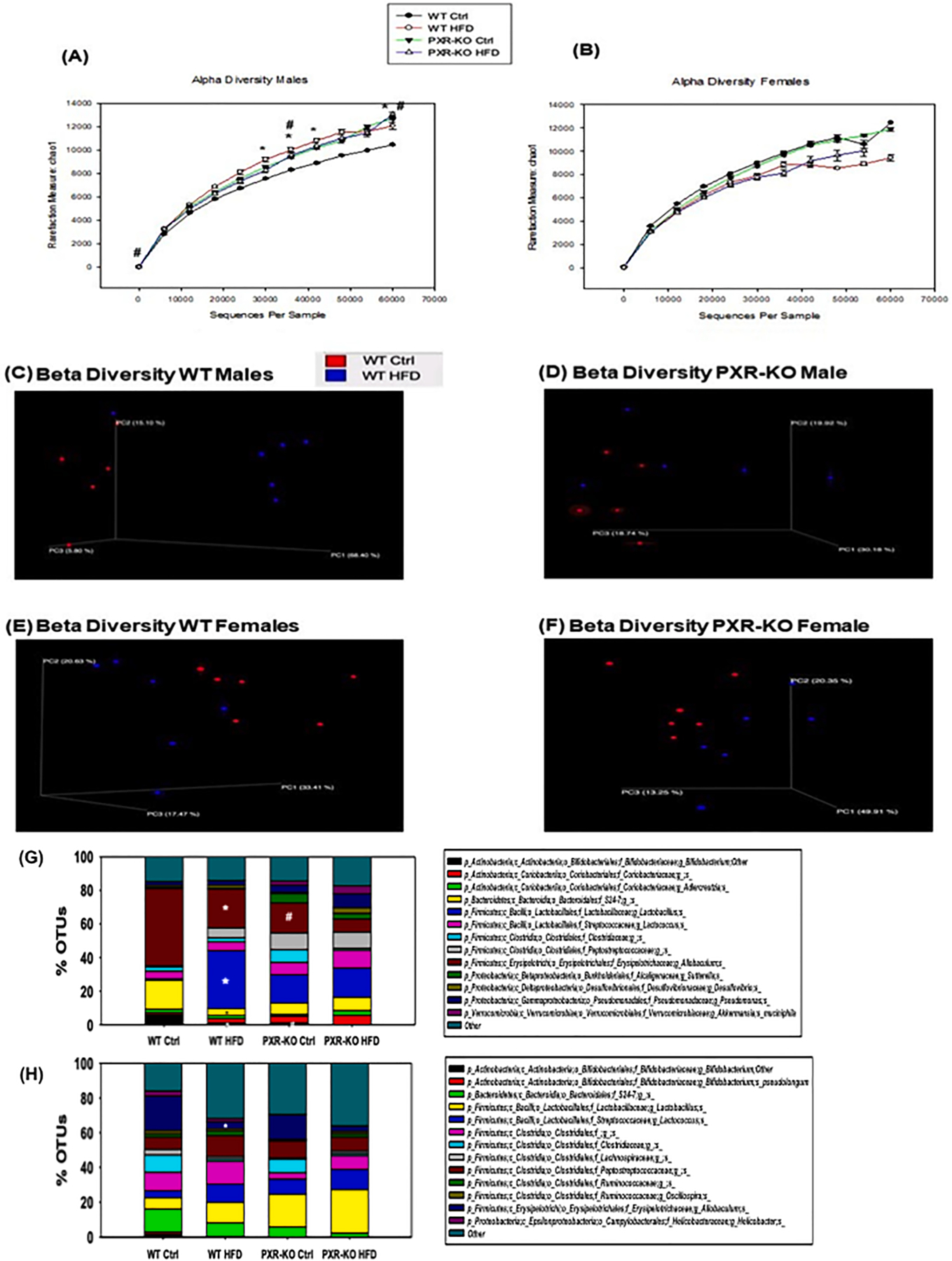
The effect of HFD on the gut microbiome from the 16S rDNA sequencing experiments. (A-B). Alpha diversities (Chao1 index) of gut microbiota in WT and PXR-KO male and female mice exposed to control diet and high fat diet (n = 6 per group). The data were analyzed using QIIME as described in the Materials and Methods section. (C-D). Beta diversities of LIC microbiota in male WT and PXR-KO mice exposed to control and high fat diet. (E-F). Beta diversity of LIC microbiota in female WT and PXR-KO mice exposed to control and high fat diet. Differentially regulated intestinal bacteria at the species level (L7) from (G) male and (H) female mice (n = 5–6 per group) (16S rDNA sequencing). Asterisks (*) represent statistically significant differences as compared with control diet and high fat diet groups; pounds (#) represent statistically significant differences as compared with WT and PXR-KO genotype groups (two-way ANOVA; statistical significance was considered at p < 0.05).
