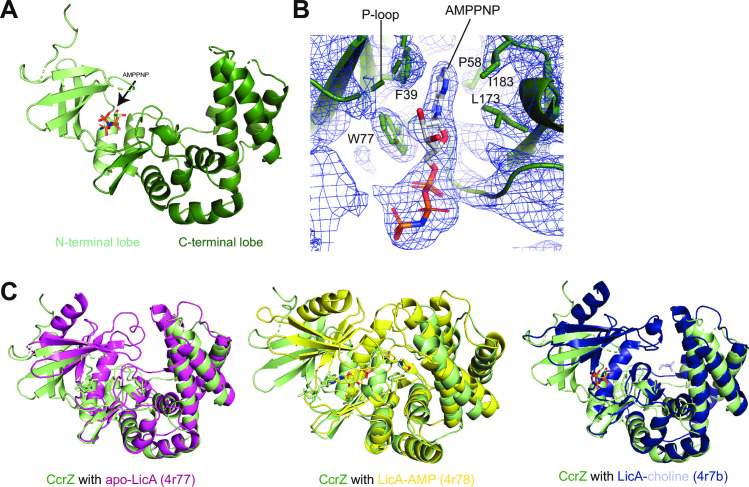
Fig 3. Structural determination of CcrZ and comparison to S. pneumoniae LicA.
(A) Structure of CcrZ bound to AMP-PNP. N-terminal lobe in light green, C-terminal lobe in dark green. (B) 2Fo-Fc map contoured at 1σ around the AMP-PNP molecule in the kinase pocket (P-loop highlighted) of the CcrZ crystal structure with aromatic and hydrophobic residues flanking the base shown as sticks. While the base occupies its pocket in the enzyme, the rest of the AMP-PNP is swung out of the active site and is directed towards the solvent. (C) CcrZ-AMP-PNP aligned with apo-LicA 4r77 (left), CcrZ-AMP-PNP aligned with LicA-AMP 4r78 (middle), CcrZ-AMP-PNP aligned with LicA-choline 4r7b (right). In all three alignments, the cleft between the N- and C-terminal lobes is wider in the CcrZ structure than in the LicA structure.