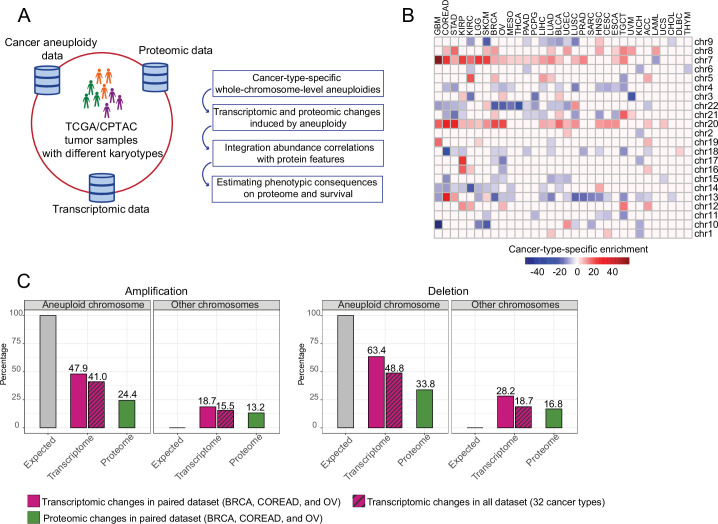
Figure 1. Transcriptomic and proteomic changes in aneuploid tumors.
(A) Data used in this study and schematic representation of the performed analyses. (B) Cancer-type-specific, whole-chromosome-level alterations across 32 cancer types. The color encodes the degree of their enrichment (standard residuals of the chi-square test multiplied by the alteration score [–1 in the case of deletion and 1 in the case of amplifications]). (C) Average percentage of differentially expressed genes or abundant proteins on aneuploid and other, non-aneuploid chromosomes (among the detected genes on the respective chromosomes).