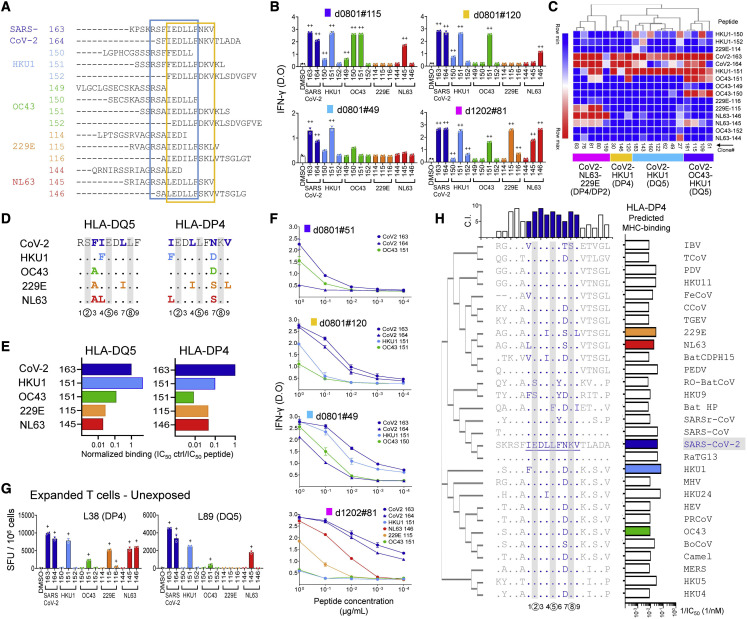
Figure 5.
Cross-reactive recognition of S811–831 homologs
(A) Sequences of SARS-CoV-2 S811–831 and corresponding HKU1, OC43, 229E, and NL63 peptides; boxes enclose the DQ5 (blue) and DP4 (yellow) 9-mer core epitopes.
(B) Responses of representative T cell clones to the peptides (IFN-γ ELISA). Bar graphs: means ± standard deviations)
(C) Hierarchical clustering of responses of 19 T cell clones to different homologs. Four major groups were defined: CoV2-OC43-HKU1/DQ5 (purple), CoV2-HKU1/DQ5 (cyan), CoV2-HKU1/DP4 (gold), and CoV2-NL63-229E/DP4-DP2 (magenta).
(D) Sequence alignment of DP4 and DQ5 core epitopes from SARS-CoV-2 and HCoVs. Numbers at bottom indicate peptide position, with T cell contacts encircled. Changes relative to SARS-CoV-2 shown in color; T cell contacts with gray bars.
(E) Normalized binding of homolog peptides relative to S811–831.
(F) Dose response of selected T cell clones to homolog peptides (IFN-γ ELISA). Symbols show means ± standard deviations.
(G) Responses of in vitro expanded cells from unexposed donors (expanded with SARS-CoV-2 peptides) to homolog peptides (IFN-γ ELISpot; +, positive response by DFR2X); relevant HLA shown in parentheses.
(H) Sequence alignment of S811–831 and positional homologs in 28 coronaviruses that sample the Orthocoronavirinae subfamily (Table S4). Complete SARS-CoV-2 sequence and differences for other viruses are shown. DP4 core epitope in blue, T cell contact positions with gray bars. Phylogenetic tree of the S proteins shown at left; conservation index (C.I.) for each position at top; predicted DP4 binding affinities at right (see also Figure S5).