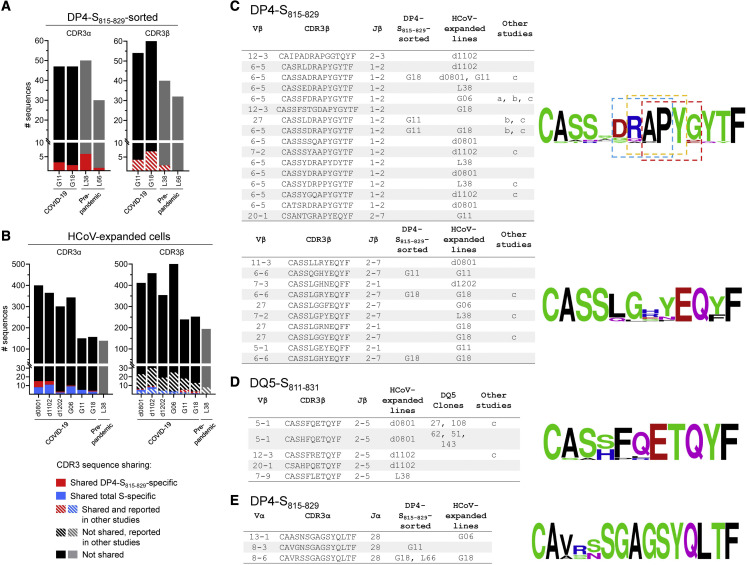
Figure 7.
Analysis of cross-reactive TCR repertoires from COVID-19 and pre-pandemic donors
(A) Summary of CDR3α and CDR3β clonotypes identified in DP4-S815-829 tetramer-sorted cells. Sequences shared among different donors shown in red, hatched lines show sequences also reported in other studies. (B) Same as previous panel, but for clonotypes identified in polyclonal lines expanded with HCoV S pool. Sequences shared among different donors shown in blue; hatched lines show sequences reported by others.
(C) Selected TCR-β DP4-specific convergence groups (CGs) identified in DP4-S815–829-tetramer sorted samples, HCoV-expanded polyclonal lines, and in other studies: a=(Nolan et al., 2020); b=(Low et al., 2021); c=(Dykema et al., 2021). Sequence logos associated with the two CGs (scores 2.8E-14 and 4.4E-14) are shown at right. Three sequence motifs identified in the first CG are boxed (Motifs: RAPY [p<0.001], APYG [p<0.001], DRAP [p<0.001]). CGs identified using GLIPH (Glanville et al., 2017).
(D) TCR-β DQ5-specific CG identified in HCoV-expanded polyclonal lines, DQ5-restricted T cell clones, and reported in other studies. CG score: 2.5E-15. CGs identified using GLIPH (Glanville et al., 2017)
(E) TCR-α DP4-specific cluster identified in DP4- S815-829-tetramer-sorted samples and HCoV-expanded polyclonal lines. Sequence cluster identified using TCRdist (Dash et al., 2017).