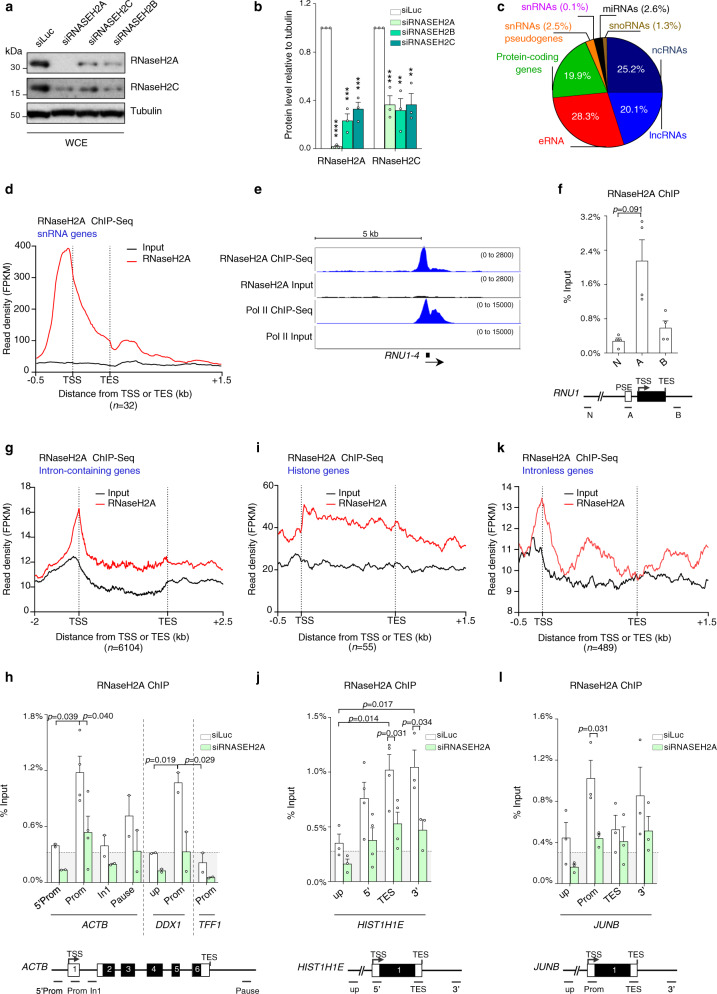
Fig. 1. RNase H2A binds active genes.
a, b Western blot of RNase H2A and H2C in the whole cell extract (WCE) in HeLa cells transfected with siLuc (white), siRNASEH2A, H2B and H2C (shades of green). Tubulin is a loading control. b Quantification of protein levels. Values are normalized to tubulin. n = 3 biologically independent experiments (means ± SEM). p-values from left to right: p < 0.0001, p = 0.0002, p = 0.0003, p = 0.001, p = 0.002, p = 0.002 (two-tailed unpaired t-test). c Distribution of RNase H2A ChIP-seq positive genes across the indicated genic compartments (n = 34,244). d Meta-analysis of read density (fragments per kilobase of transcript per million mapped reads, FPKM) for RNase H2A ChIP-seq on snRNA genes across −0.5 kb genomic region flanking transcription start site (TSS) and +1.5 kb transcription termination site (TES) in HeLa cells. All subsequent images show one representative replicate out of 2 with RNase H2A signal in red and input in black. Number of genes analyzed are shown in brackets. e RNase H2A and Pol II ChIP-seq profiles of RNU1-4 in HeLa cells. Numbers in brackets indicate the viewing range (FPKM). f RNase H2A ChIP-qPCR for RNU1 in HeLa cells. Values are expressed as percentage of input. n = 4 biologically independent experiments (means ± SEM). p-values were calculated using two-tailed unpaired t-test. All subsequent gene diagrams depict snRNA coding region or protein-coding gene exons (black), UTRs (white), introns (lines), TSS, TES and qPCR amplicons (below the diagram). PSE is proximal sequence element. g, i, k Meta-analysis of read density (FPKM) for RNase H2A ChIP-seq on different categories of genes in HeLa cells, with the window size of the presented plots adjusted to the gene size. Data are represented as in d. h, j, l RNase H2A ChIP-qPCR in HeLa cells transfected with siLuc (white bars) and siRNASEH2A (green bars) for different gene categories. Values represent percentage of input (means ± SEM). p-values were calculated using two-tailed unpaired t-test. h Intron-containing genes (ACTB, DDX1 and TFF1). n = 2 (n = 4 for ACTB Prom) biologically independent experiments. j Histone HIST1H1E gene. n = 3 (n = 4 for 5’ and TES) biologically independent experiments. l Intronless JUNB gene. n = 3 biologically independent experiments. Horizontal dotted line indicates background signal. Source data are provided as a Source data file.