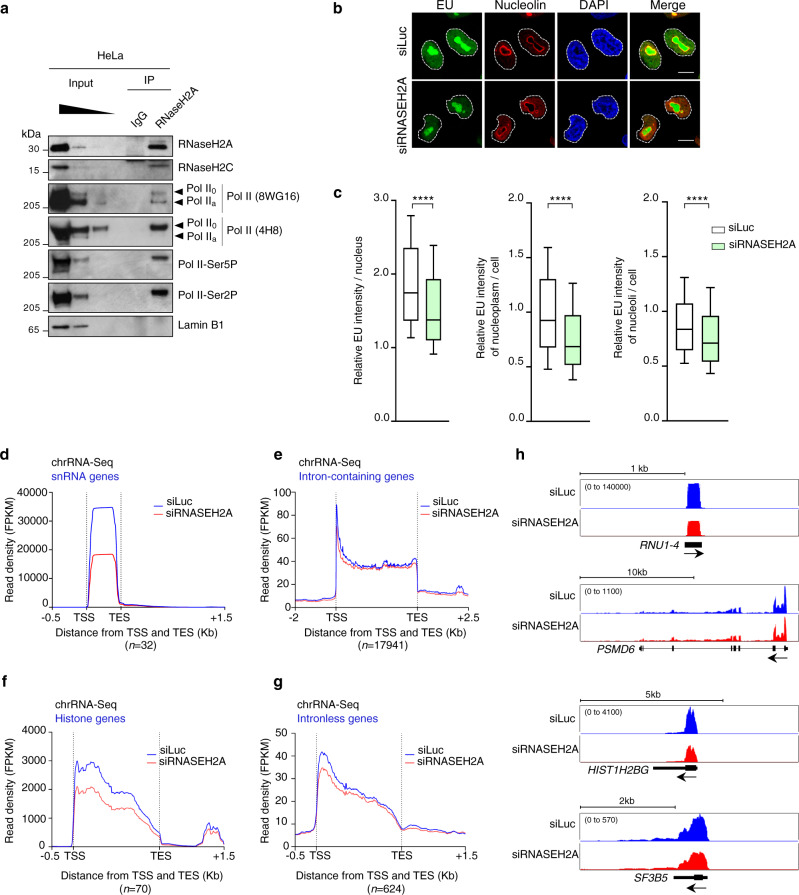
Fig. 3. RNase H2A depletion affects transcription.
a Western blot of endogenous RNase H2A IP from HeLa WCE probed with the indicated antibodies. Arrows indicate hypo- (IIa) and hyper-phosphorylated (IIo) Pol II. IgG and Lamin B1 are negative controls. Representative blots from n = 2 biologically independent experiments. b IF analysis of EU incorporation in HeLa cells transfected with siLuc or siRNASEH2A. EU (green), nucleolin (red), DAPI (blue), Nucleolin + EU (merge); scale bars: 10 µm. Representative images from n = 2 biologically independent experiments. c Quantification of EU intensity per nucleus (left panel), nucleoplasm/cell (nucleus without nucleolus; middle panel) or nucleoli/cell (right panel) in HeLa cells transfected with siLuc (white) or siRNASEH2A (green). EU intensity is normalized to the average nucleoplasmic intensity in siLuc condition. Boxplot settings are: box: 25–75 percentile range; whiskers: 10–90 percentile range; horizontal bars: median; outliers not displayed. >20,000 nuclei were quantified per condition. Representative data from n = 3 biologically independent experiments. ****p < 0.0001 (two-tailed unpaired t-test). d–g Meta-analysis for chrRNA-Seq on indicated categories of genes in HeLa cells following RNase H2A depletion with the window size of the presented plots adjusted to the gene size. siLuc (blue) and siRNASEH2A (red) tracks show the average signal of two replicates. h ChrRNA-seq profiles of RNU1-4, PMSD6, HIST1H2BG and SF3B5 genes in HeLa cells treated as in d–g. Source data are provided as a Source data file.