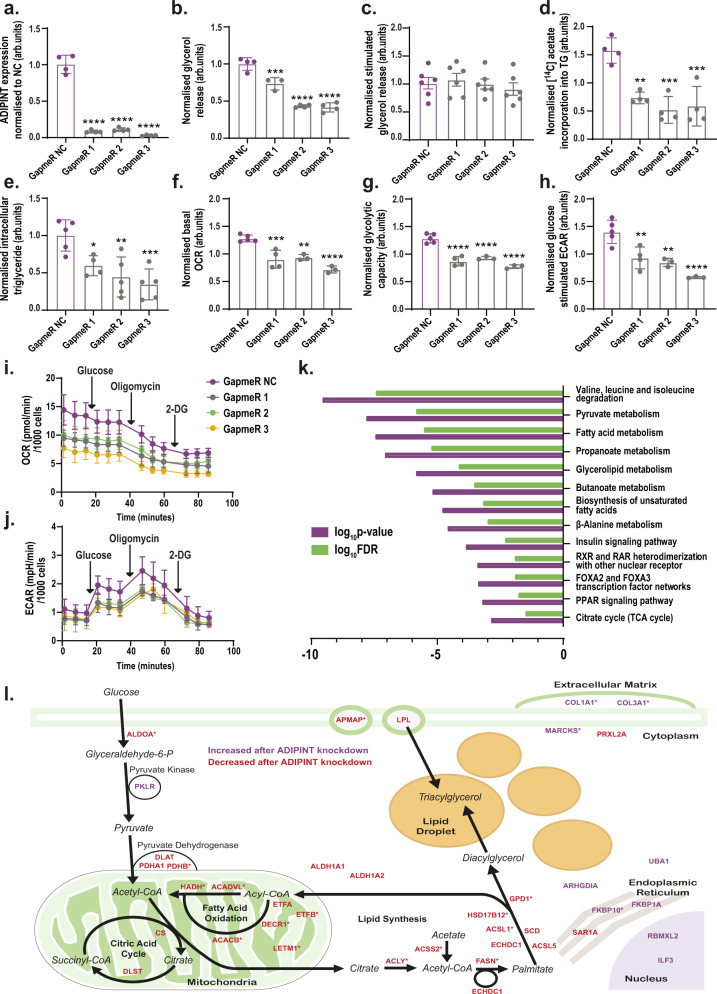
Fig. 2. Knockdown of ADIPINT perturbs lipid metabolism and bioenergetics.
a ADIPINT expression (n = 4) b glycerol release (measure of basal lipolysis) (n = 3,4) c isoprenaline stimulated glycerol release (measure of stimulated lipolysis) (n = 6) d insulin stimulated [14C] acetate incorporation into intracellular lipid (measure of lipid synthesis) (n = 4) e intracellular triglyceride (TG) content (n = 4,5) f basal oxygen consumption rate (OCR) (n = 3–5) g glycolytic capacity (n = 3–5) and h glucose-stimulated extracellular acidification rate (ECAR) (n = 3–5) in differentiated hADSC transfected with three ADIPINT targeting GapmeRs (1–3) or non-targeting control GapmeR (NC). Each data point represents an individual experiment. Data was normalized to the mean of each replicate experiment apart from d mean normalized to GapmeR NC (-insulin). ATP linked OCR was determined after oligomycin administration. ECAR determined after glucose treatment. i The OCR and j ECAR measured at basal and then after glucose, oligomycin and 2-deoxyglucose (2-DG) treatment over the time course as indicated, n = 5 for GapmeR NC, n = 4 for GapmeR 1 and n = 3 for GapmeR 2 and 3 independent experiments. a–h Each data point represents an independent experiment. One-way ANOVA with Dunnett’s post-hoc test was used to assess significance between GapmeR NC and GapmeRs 1–3. k Pathways enriched by genes downregulated after ADIPINT knockdown by each GapmeR independently, compared to GapmeR NC. l A schematic diagram of all 38 proteins significantly altered independently by all three GapmeRs targeting ADIPINT. Proteins are displayed based on the associated metabolic pathway and localization according to www.genecards.org. Proteins downregulated following ADIPINT knockdown are in red font and upregulated are in purple font. Proteins marked with * were not significantly regulated at the RNA level. If no metabolic pathway was listed for the given protein, then the protein was placed on the diagram based on the given subcellular localization. Metabolites were not measured. Data are presented as mean values + /− SD for n < 6 and SEM for n ≥ 6. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Source data are provided as a Source Data file.