Figure 1.
Architecture of the CCoV-HuPn-2018 infection machinery
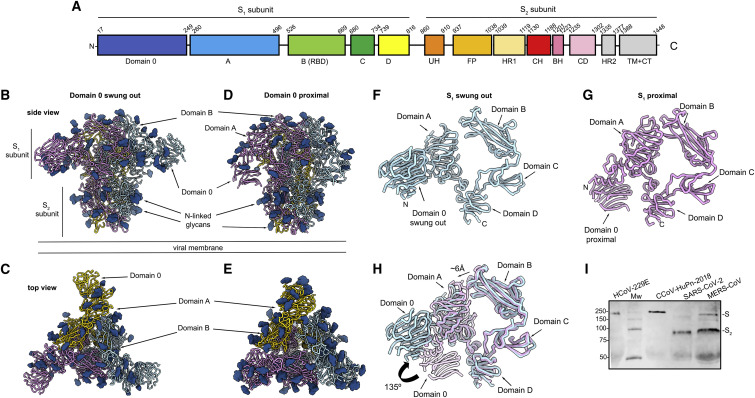
(A) Schematic diagram of the S glycoprotein organization. UH, upstream helix; FP, fusion peptide; HR1, heptad-repeat 1; CH, central helix; BH, β-hairpin; CD, connector domain; HR2, heptad-repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. Gray boxes denote regions unresolved in the reconstruction (HR2) and regions that were not part of the construct (TM and CT), respectively.
(B and C) Cryo-EM structure of the CCoV-HuPn-2018 S ectodomain trimer (with each domain 0 swung out) viewed along two orthogonal orientations.
(D and E) Cryo-EM structure of the CCoV-HuPn-2018 S ectodomain trimer (with each domain 0 pointing “proximal” toward the viral membrane) viewed along two orthogonal orientations.
(F and G) Ribbon diagram of the CCoV-HuPn-2018 S1 subunit with domain 0 in the swung out (F) and proximal (G) conformations.
(H) Superimposition of the S1 subunit from the structures with domain 0 in the swung out (light blue) and proximal (pink) conformations showing the A domain moving away from the B domain of the same protomer and rotation of domain 0.
Renderings in (B–H) use composite models obtained from the global and local refinements for each conformation.
(I) Western blot of VSV pseudotyped particles harboring HCoV-229E S, CCoV-HuPn-2018 S, SARS-CoV-2 S, and MERS-CoV S. MW, molecular weight ladder. Full-length S and S2 subunit bands are indicated on the right-hand side of the blot.
See also Figures S1 and S2 and Tables S1 and S2.