Figure 1:
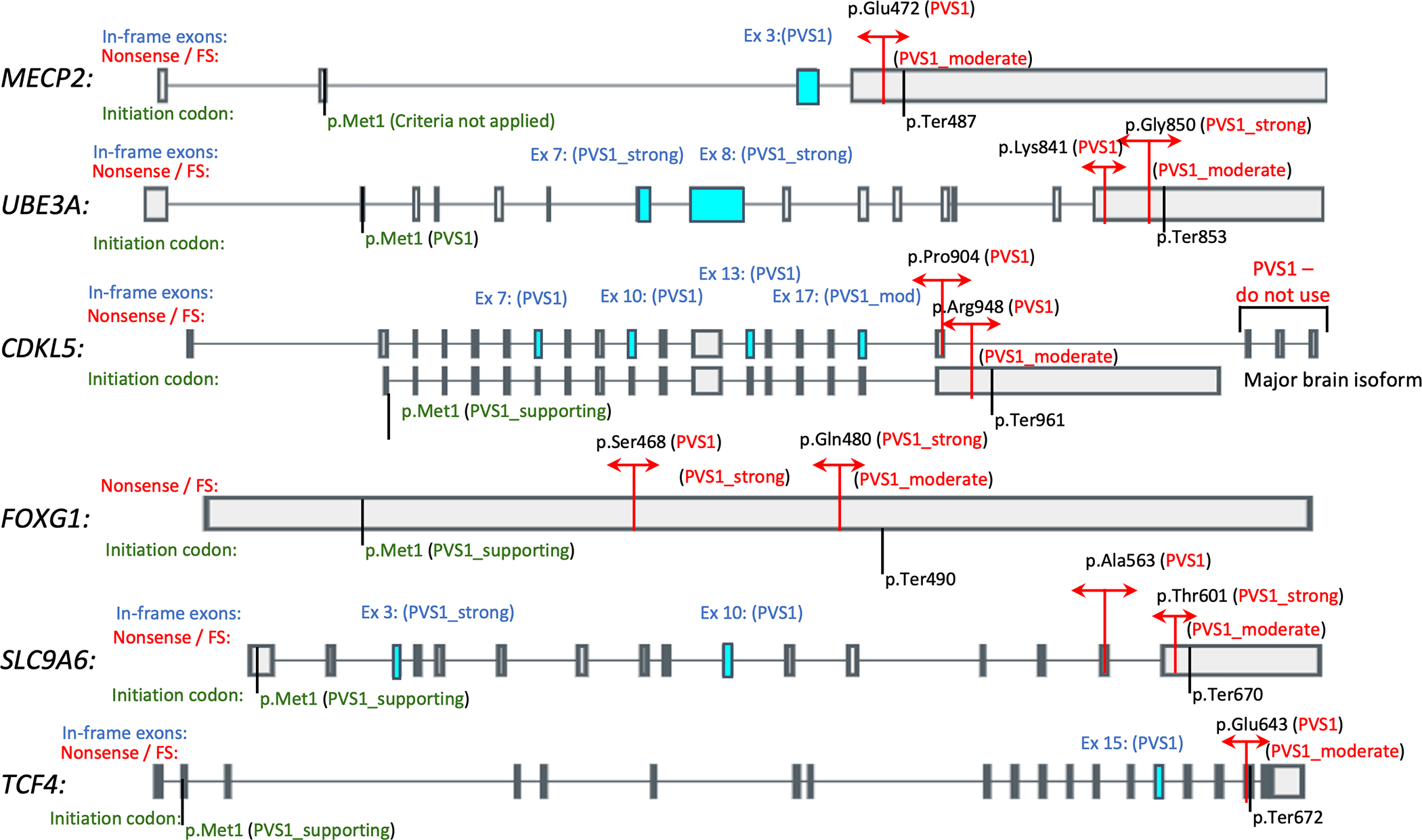
Diagrammatic representation of PVS1 cutoffs for each gene. Cutoffs for nonsense and frameshift variants are indicated in red. In-frame exons for each gene are indicated in blue with the respective PVS1 strengths indicated for deletion/skipping of these exons also in blue. PVS1 strengths for variants affecting the initiation codon are indicated in green. MECP2: NM_004992.3 / ENST00000303391.10; UBE3A: NM_130838.4 / ENST00000438097.6; CDKL5: NM_003159.2 / ENST00000379996.7 (top), NM_001323289.2 / ENST00000623535.1 (bottom); FOXG1: NM_005249.4 / ENST00000313071.6; SLC9A6: NM_006359.2 / ENST00000370698.7; TCF4: NM_001083962.1 / ENST00000354452.7
