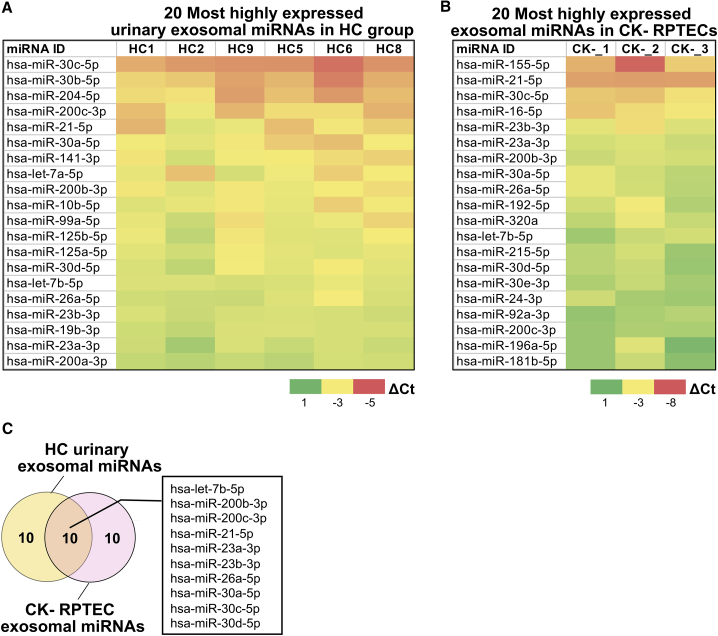
Figure 1.
Urinary exosomal miRNAs that are potentially derived from RPTECs
(A) Top 20 most abundant exosomal miRNAs from healthy control (HC) group (n = 6) and (B) non-stimulated RPTECs (CK−) (n = 3). Heatmap colors indicate the delta Ct (ΔCt) value of each target miRNA. Red indicates lower ΔCt (higher gene expression) and green indicates higher ΔCt (lower gene expression). (C) Overlap between the 20 most abundant exosomal miRNAs derived from CK− RPTECs and that from urine of HC group.