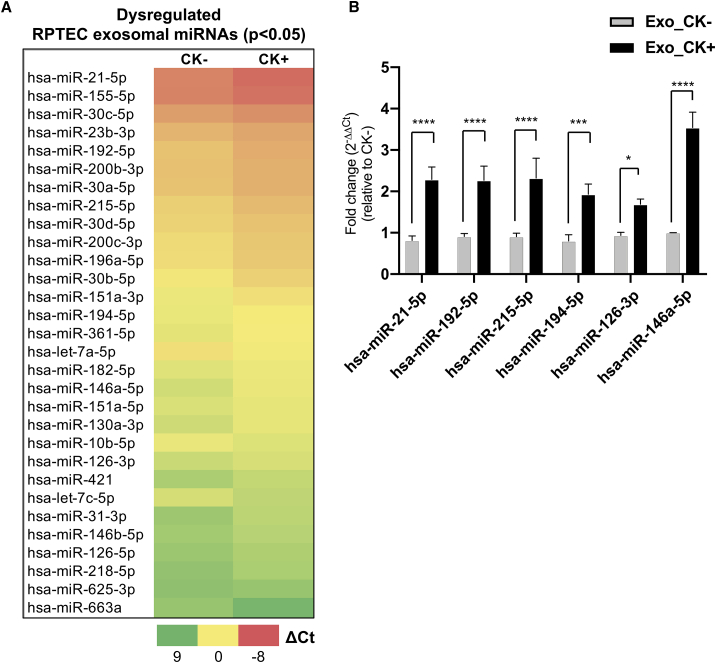
Figure 3.
Dysregulated exosomal miRNAs from cytokine-treated RPTECs as potential indicators of inflammatory kidney disease
(A) Identification of dysregulated exosomal miRNAs (p < 0.05) derived from non-stimulated (CK−) and cytokine-stimulated (CK+) RPTECs (see section “materials and methods”). Heatmap represents the average ΔCt values of each miRNA in CK− or CK+. Red indicates lower ΔCt (higher gene expression) and green indicates higher ΔCt (lower gene expression). Student's t test was employed for statistical analysis using the ΔCt values obtained from three independent experiments. (B) Validation of six differentially abundant candidate exosomal miRNAs. The relative fold change (FC) of exosomal miRNA expression in cytokine-stimulated condition (Exo_CK+) relative to non-stimulated condition (Exo_CK−) was analyzed using 2−ΔΔCt method. Bar graph represents mean ± SEM of five independent experiments. Two-way ANOVA with Benjamini and Hochberg multiple comparisons was performed for statistical analysis using (GraphPad Prism, 8.0.1). ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗p < 0.05.