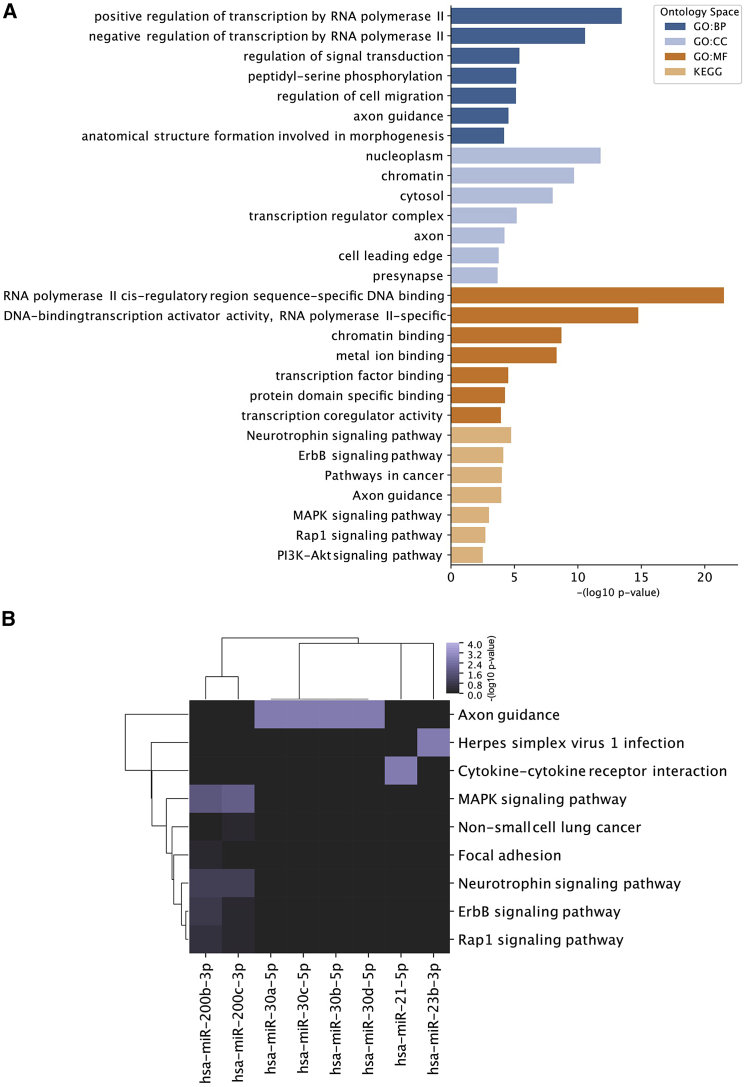
Figure 4.
Pathway analysis of dysregulated and highly abundant exosomal miRNAs
(A) Gene Ontology analysis of the target space derived from 12 dysregulated and highly abundant exosomal miRNAs (hsa-miR-155-5p, -192-5p, -196a-5p, 200b/c-3p, -21-5p, 215-5p, 23b-3p, and -30a/b/c/d-5p) was performed for the ontology spaces (GO:BP, GO:CC, GO:MF, and KEGG) and the top seven enriched pathways are represented by the ranked negative log10 p value for each ontology spaces. (B) KEGG pathway analysis of the target space (retrieved from the DIANA microT-CDS prediction tool [score >0.9]) for each of the 12 most significantly dysregulated and highly abundant exosomal miRNAs from cytokine-stimulated RPTECs. Enriched pathways are represented and ranked by the negative log10 p value.