Figure 6.
Validation of dysregulated exosomal mt-tRNAs, mt-tRNAHis, mt-tRNALeu2, mt-tRNAVal, mt-tRNAPhe, and mt-tRNALeu1 derived from an RPTEC model system of renal inflammation
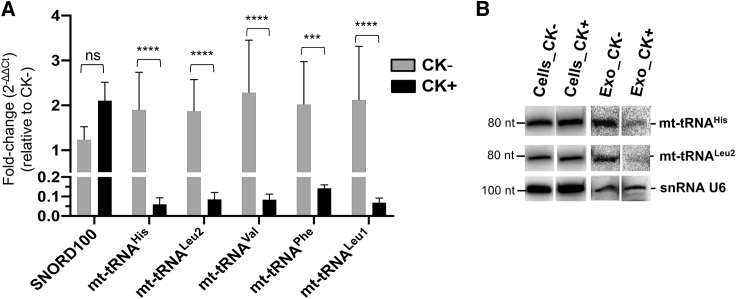
(A) RT-qPCR analysis of total exosomal RNA extracts from either non-stimulated (CK−) or cytokine-stimulated (CK+) RPTECs (see section “materials and methods”). The FC expression of each candidate, relative to CK− samples, was analyzed by the 2−ΔΔCt method. Data indicate mean ± SEM of four independent experiments. Statistical analysis of ΔCt values was performed using two-way ANOVA with multiple comparison test and Bonferroni correction (GraphPad Prism 8.0.1). ∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; and ns, not significant. (B) Northern blot analysis of mt-tRNAHis and mt-tRNALeu2 (see section “materials and methods”). Total RNA extracts were derived from either RPTEC exosomes (Exo) or cells under non-stimulated (CK−) or cytokine-stimulated (CK+) condition.