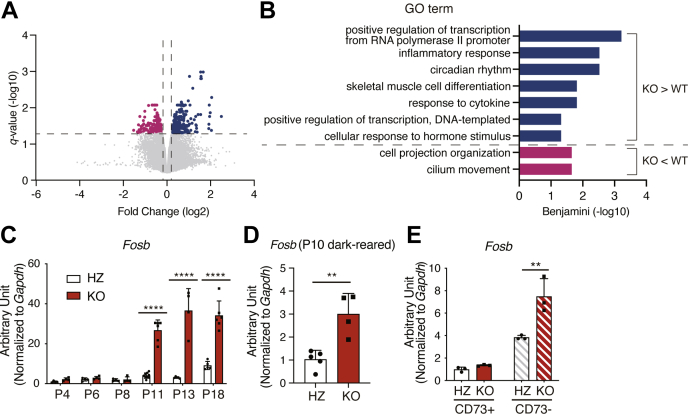
Figure 4.
Transcriptomic analysis of Lpcat1 knockout (KO) retina.A, volcano plot of differentially expressed genes (DEGs) between postnatal day 14 (P14) Lpcat1 wildtype (WT) and KO retina. DEGs (fold change >1.2 or <−1.2, q-value < 0.05) increased in Lpcat1 KO are highlighted in blue, and DEGs decreased in Lpcat1 KO are highlighted in magenta (n = 4 for each group). B, functional annotation of upregulated or downregulated genes (shown in blue or magenta in A) in Lpcat1 KO retina. C–E, the mRNA expression of Fosb during neonatal development (C, n = 4 for HZ and n = 9 for KO), in P10 dark-reared littermates (D, n = 4 for HZ and n = 5 for KO), and in CD73-positive (photoreceptor cells) or CD73-negative cells (E, n = 3 for each group) of Lpcat1 heterozygous (HZ) and KO retina. The expression level was normalized by glyceraldehyde 3-phosphate dehydrogenase (Gapdh). Data are shown as mean + SD of independent experiments. C and E, significance is based on two-way ANOVA followed by Bonferroni’s multiple comparisons test (∗∗p < 0.01, ∗∗∗∗p < 0.0001). D, significance is based on unpaired t test (∗∗p < 0.01).