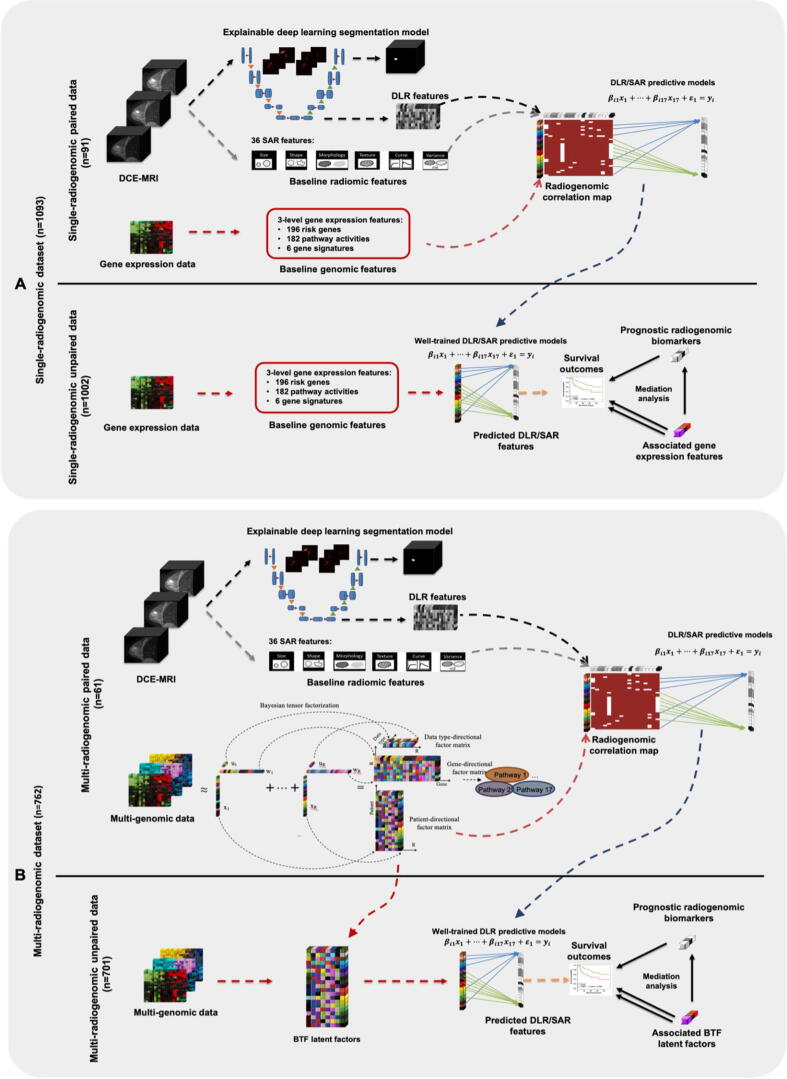
Fig. 1.
The overall workflow of this study. A DL model (3DU-net) was built, trained, and validated to segment the tumor region from the raw three-dimensional DCE-MRI image. After the 3DU-net were well-trained, DL-based radiomic (DLR) features were extracted from the last hidden layer in the encoding phase of the model. Gradient-based saliency maps were generated to show the importance of each input pixel to the 3DU-net of making its segmentation decision. A: Single-radiogenomic stage. In this stage, we first focus on the paired data (top panel of A). Three-level gene expression features (197 breast cancer risk gene expressions, 182 KEGG pathway activities, and 6 well-established breast cancer gene signatures) are generated. Then, lasso models are built to predict each DLR feature and semi-auto radiomic (SAR) feature using these three-level gene expression features. After the predictive lasso models are well-trained and validated, we turn to the unpaired data (bottom panel of A). We generate the same three-level gene expression features using the unpaired data, then we apply the well-trained lasso models to get the predicted DLR and SAR features. In this way, we could generate the DLR and SAR features for the 1002 patients without medical images. Then, we performed survival analysis on the predicted DLR and SAR features. The significant ones are the identified prognostic radiogenomic biomarkers. Mediation analysis is then performed on these identified radiogenomic biomarkers to check the potential biological mechanisms of them. B: Multi-radiogenomic stage. In this stage, similar procedures of the single-radiogenomic stage are performed. We first focus on the paired data (top panel of B). We perform Bayesian tensor factorization (BTF) on the multi-genomic data tensor to extract 17 BTF features. We also run gene set enrichment analysis (GSEA) to identify the key biological pathway of each BTF feature. These key pathways could explain the key functions of the identified multi-genomic BTF features. Then we train lasso models to utilize these 17 BTF features for predicting the DLR and SAR features. After the lasso models are well-trained and well-validated, we turn to the unpaired data (bottom panel of B). We obtain the BTF features using the multi-genomic data, then we apply the well-trained lasso models in the previous step to get the predicted DLR and SAR features. In this way, we could get the DLR and SAR features for the 701 patients without medical images. Then, we perform survival analysis on the predicted DLR and SAR features. The significant ones are the identified radiogenomic biomarkers. Mediation analysis is then performed on each of these identified radiogenomic biomarkers to check the potential biological mechanisms of them.