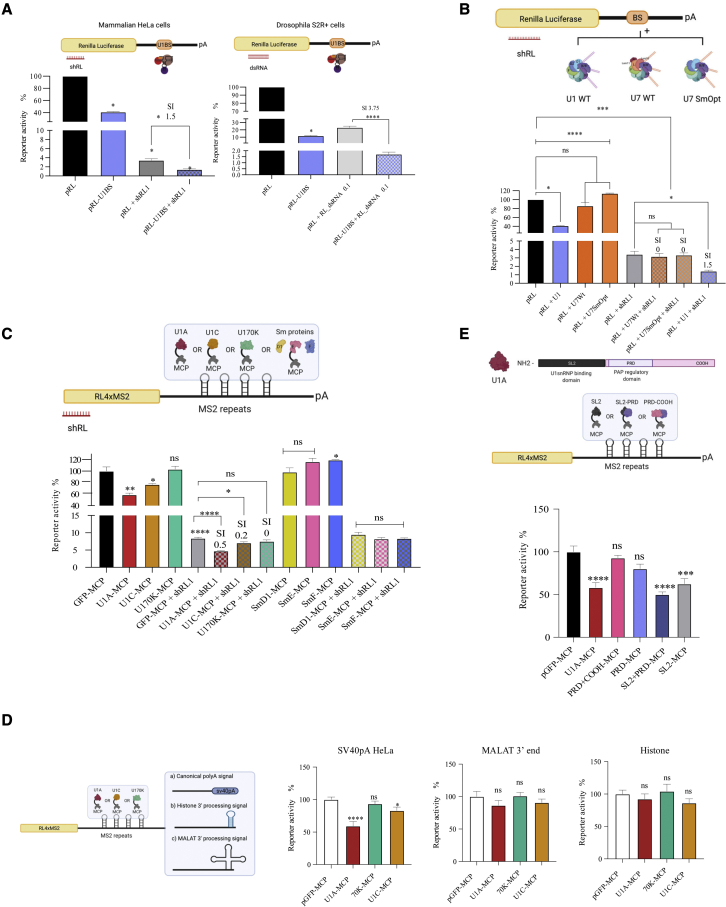
Figure 1.
Inhibition of gene expression by RNAi, U1i, or U1 snRNP proteins
(A) HeLa or S2R + Drosophila cells were transfected with a plasmid expressing an RL transcript with a wild-type (WT) (pRL-U1BS) or a mutated (pRL) binding site for endogenous U1 snRNP and/or a plasmid expressing a shRNA (HeLa) or 0.1 ng of dsRNAs (S2R+) against RL. (B) HeLa cells were co-transfected with plasmids expressing RL, plasmids expressing U1WT, U7WT, or U7SmOpt snRNAs, which target RL 3′ UTR (BS) and/or plasmids expressing a shRNA against RL. (C) Mammalian cells were co-transfected with a plasmid expressing an RL transcript with 4 MS2-binding sites, plasmids expressing the indicated MCP fused proteins, and/or plasmids expressing a shRNA against RL. (D) As in (C), but the RL 4xMS2 plasmid contains a canonical, a histone, or a MALAT1 3′-end processing signal. Each plasmid was co-transfected with plasmids expressing MCP fused to U1 snRNP-specific proteins. (E) As in (C), but the RL 4xMS2 plasmid was co-transfected with plasmids expressing MCP fused to U1A or U1A-truncated fragments. In all (A–E) cases, a plasmid expressing firefly luciferase (luc) was also co-transfected as a transfection control. Two days after transfection, RL and luc were quantified and the activity of RL was normalized with luc to calculate the percentage of reporter activity. All experiments were performed at least three times in triplicate. Error bars show standard error of the mean (SEM). The synergy index (SI) was calculated as described.15 Either two-tailed Student’s t test or one-way ANOVA was employed to compare two or more independent groups respectively. Results are indicated as ns, non-significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001.