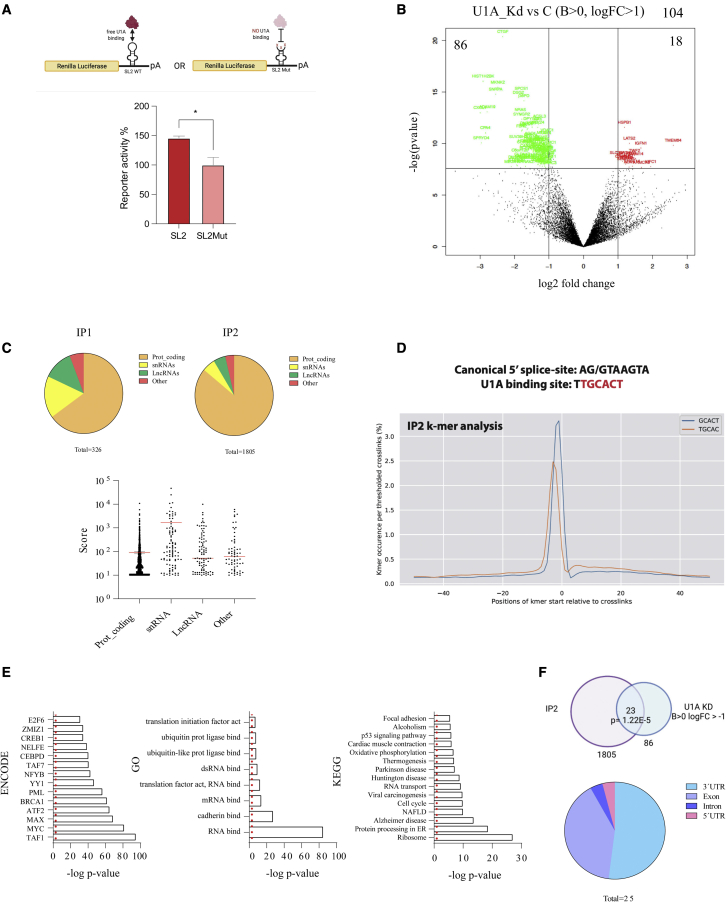
Figure 2.
Free U1A binds target genes and is a positive regulator of gene expression
(A) HeLa cells were transfected with plasmids expressing an RL transcript with a WT or a mutant SL2 sequence in the 3′ UTR (schematized in the upper part of the figure) and luc as a transfection control. Two days after transfection, RL and luc were quantified and the activity of RL was normalized with luc and used to calculate the percentage of reporter activity. The experiment was performed at least three times in triplicate. Error bars show SEM. Two-tailed Student’s t test was employed; ∗p < 0.05. (B) Volcano plot showing genes deregulated (B > 0, logFC>1) after analysis of 3′-end RNA sequencing (RNA-seq) of cells transfected with control or U1A targeting siRNAs. The experiment was performed in triplicate. Downregulated genes are in green and upregulated genes are in red. (C) Pie chart showing the RNA subtypes co-immunoprecipitated with 3xFLAG-U1A in IP1 and IP2 iCLIP data (C, top); graph with the iCLIP peak score for each RNA identified in IP2 (n = 1815) classified by RNA subtypes (C, bottom). (D) Graph with the K-mer sequence enriched in IP2 iCLIP data, which shows U1A consensus sequence. (E) Bar plots with the Enrich R analyses for ENCODE transcription factors, GO terms, and KEGG pathways with the list of U1A-bound genes from IP2 iCLIP data. (F) Overlap between U1A-bound genes from IP2 iCLIP data and genes downregulated after U1A knockdown from 3′ -end RNA-seq data. The result from hypergeometric enrichment analysis is shown. Pie chart showing the distribution of U1A iCLIP peaks (n = 25) within the 23 genes identified.