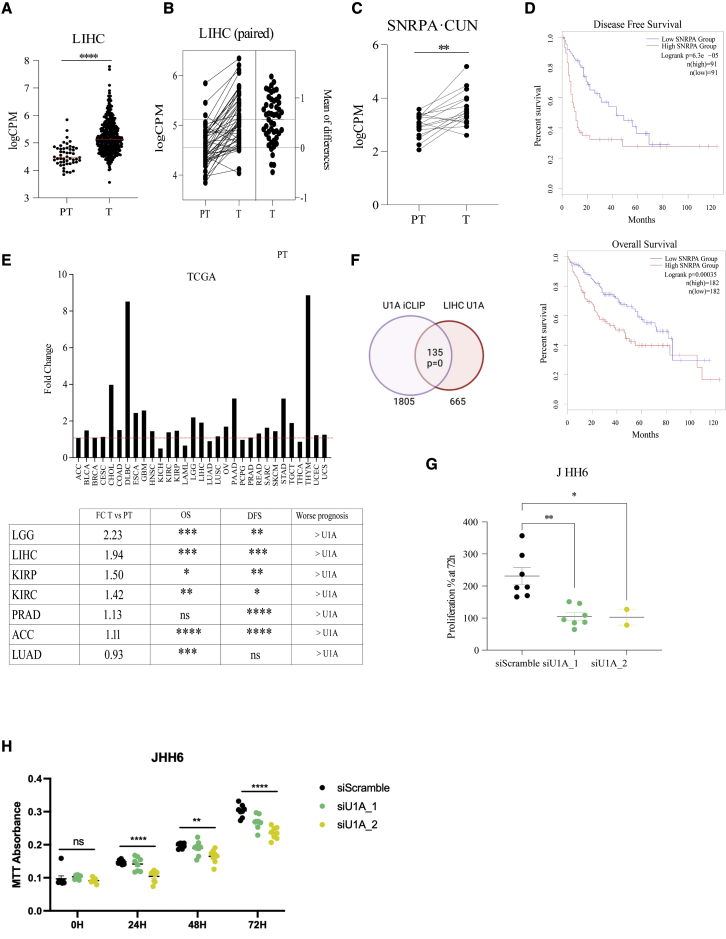
Figure 4.
U1A mRNA levels associate with cancer prognosis and cell growth
(A–C) U1A mRNA levels were evaluated in data from TCGA (A) or in paired samples from TCGA (B) or our own cohort of patients (C). (D) Disease-free survival (DFS) and overall survival (OS) were evaluated with the TCGA cohort of LIHC using GEPIA. (E) Top: fold change (FC) increase of U1A mRNA levels in tumor (T) versus peritumor (PT) samples from the TCGA cohort of indicated tumors. Bottom: TCGA patients were divided in two groups of equal size based on the U1A mRNA level in the tumor (high or low) and DFS and OS were calculated for the indicated tumors. (F) Overlap of U1A-bound genes from U1A IP2 iCLIP data and genes whose expression positively correlates with U1A mRNA levels (R > 0.5) in TCGA LIHC data. The result from hypergeometric enrichment analysis is shown. (G) JHH6 cells were transfected with control or two independent U1A-targeting siRNAs and cell number was quantified 3 days later. The percentage of proliferation, shown in the y axis, was calculated by dividing the final cell number by the number of cells seeded prior to transfection. Proliferation percentage at 72 h post transfection is plotted as dots representing each replicate. Error bars show SEM. Two-tailed Student’s t test was employed. ∗ p < 0.05 and ∗∗p < 0.01. (H) JHH6 cells were transfected with control or two independent U1A-targeting siRNAs and cell proliferation was assessed through an MTT assay. MTT absorbance is plotted as dots representing each replicate. Error bars show SEM. One-way ANOVA was employed to compare group means: ns, not significant; ∗∗p < 0.01; ∗∗∗∗p < 0.0001.