Figure 4.
Mass spectrometry results and mechanism
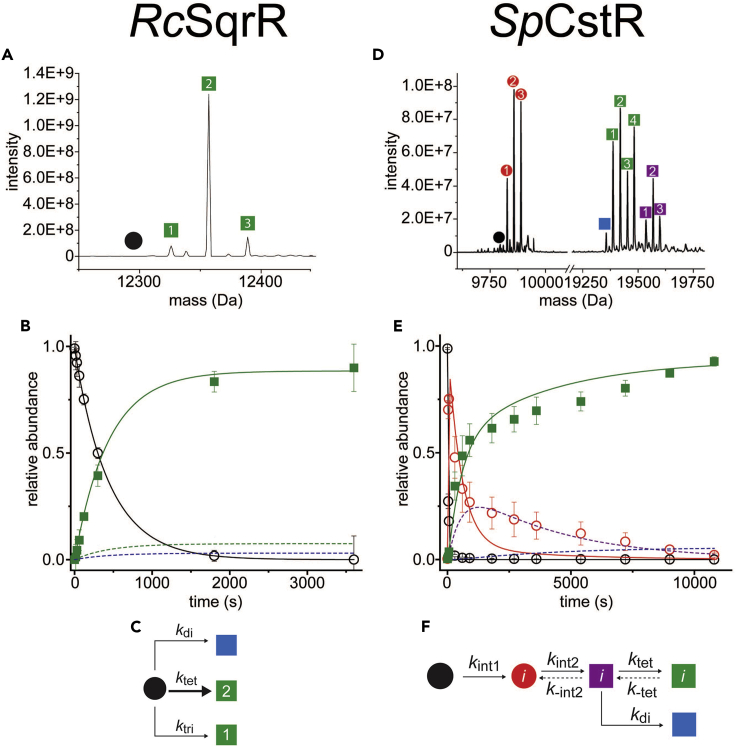
(A) Representative regions of the ESI-MS data obtained for a reaction of GSSH with RcSqrR. The symbols correspond to the reduced and IAM-capped (black) and tetrasulfide (green) monomeric species. No crosslinked dimer species are observed here since the cysteines that form the tetrasulfide linkage are derived from the same protomer (Capdevila et al., 2021).
(B) Relative abundance of the indicated species as a function of pulse time t fitted to the kinetic mechanism in panel C, represented by the continuous and dashed lines. Datapoints for the dashed lines corresponding to disulfide (red) and trisulfide (green) were omitted to improve the clarity.
(C) A minimal kinetic model with respect to the protein concentration without an intermediate step. Major species formed (tetrasulfide) is represented with a bold arrow. The two minor species (disulfide and trisulfide) were fitted to parallel reactions from the same starting material.
(D) Representative regions of the ESI-MS data obtained for a 10 min reaction of CysSSH with SpCstR. In each case, the monomer region is shown at left, and the crosslinked dimer region is shown at right. The symbols correspond to the following: capped monomer (black), polysulfidated monomer (red), closed/open (purple), di/di (blue), and closed/closed (green).
(E) Global analysis of the kinetic profiles for SpCstR using the generalized kinetic scheme and fitted to the minimal bifurcated model shown in panel F, represented by continuous and dashed lines. Datapoints for the dashed lines corresponding to closed/open (purple) and di/di (blue) were omitted to improve the clarity.
(F) Generalized kinetic scheme used to analyze the reaction profiles obtained with persulfide donors. Continuous lines represent the processes that were determined in all profiles, whereas dashed lines represent the processes that could only be determined for some profiles (Fakhoury et al., 2021).