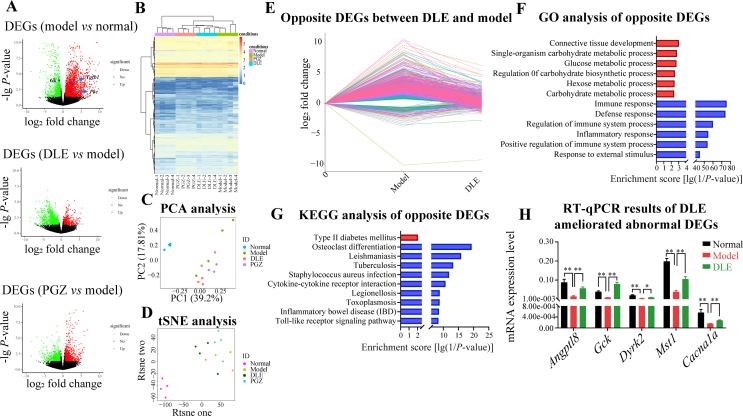
Figure 3.
DLE affected glucose and lipid metabolic pathways and partly reversed diabetes-induced transcriptional changes in vivo. (A) Volcano plots of DEGs among different comparisons (upregulation in red and downregulation in green). Each comparison of DEGs was determined by comparing model vs. normal, DLE vs. model, and PGZ vs. model using fold change >1.5 and P-value < 0.05 as the cut-off. (B) Hierarchical clustering, (C) PCA, and (D) tSNE analysis of all groups was performed using the normalized RNA-seq read counts that corresponded to all DEG unions as the input. (E) Line chart of DLE improved 1004 DEGs in db/db mice (opposite DEGs between DLE and model group). (F) The GO annotated biological processes enriched by the DLE improved 1004 DEGs. Among them, 101 were upregulated (red) and 903 were downregulated (blue). (G) Enriched KEGG pathways in the 101 DLE-upregulated genes (red) and 903 DLE-downregulated genes (blue). (H) RT-qPCR of representative genes involved in glucose metabolism pathways (n ≥ 3). The data are shown as the mean ± SD. Statistical analyses were conducted using unpaired Student's t-tests. ∗P < 0.05, ∗∗P < 0.01 as indicated in the figure. DEG, differentially expressed genes; PCA, principal component analysis; tSNE, t-distributed stochastic neighbor embedding; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; RT-qPCR, quantitative reverse transcription polymerase chain reaction.