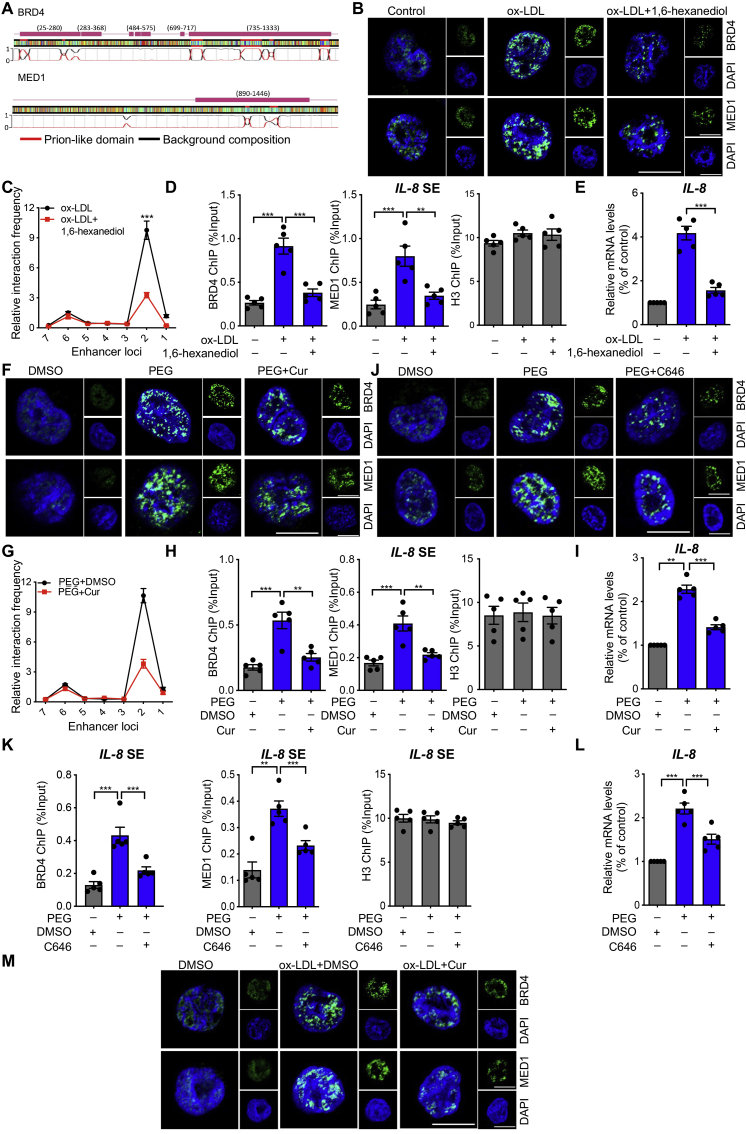
Figure 6.
Cur regulates BRD4-dependent SE associated with phase separation on inflammatory genes. (A) A color-coded schematic representation of the aligned amino acid sequences and corresponding prion-like domain disorder propensity plots for BRD4 and MED1 generated using the prion-like amino acid composition pool. The gray line indicates the low complexity region. (B) Immunofluorescence analysis of the puncta of BRD4 (green) and MED1 (green) in ox-LDL-treated macrophages exposed to 3% 1,6-hexanediol for 15 s. Scale bar = 20 μm. (C) 3C-qPCR analysis of the long–distance interactions between the IL-8 promoter and seven enhancer loci (the positions of the different loci are described in Fig. 5A) in FCs treated with 1,6-hexanediol (x-axis means seven enhancer loci of IL-8 promoter). (D) ChIP analysis of the enrichment of BRD4 and MED1 at the SE regions of IL-8 in ox-LDL-treated macrophages combined with 3% 1,6-hexanediol, as well as H3. (E) Relative mRNA level of IL-8 in ox-LDL-treated macrophages exposed to 3% 1,6-hexanediol. (F–L) Macrophages were treated with PEG along with Cur or C646. We detected the puncta of BRD4 (green) and MED1 (green) by immunofluorescence (F, J), the long–distance interactions between IL-8 promoter and seven enhancer loci in macrophages with different treatments by 3C-qPCR (x-axis means seven enhancer loci of IL-8 promoter) (G), the enrichment of BRD4 and MED1 at the SE regions of IL-8 by ChIP assay (H, K), and the mRNA level of IL-8 by qRT-PCR (I, L). Scale bar = 20 μm. (M) Immunofluorescence analysis of the puncta of BRD4 (green) and MED1 (green) in ox-LDL-treated macrophages combined with Cur. Scale bar = 20 μm. Data are expressed as mean ± SEM, n = 5. ∗∗P < 0.01, ∗∗∗P < 0.001.