Abstract
Sequence variants affecting blood lipids and coronary artery disease (CAD) may enhance understanding of the atherogenicity of lipid fractions. Using a large resource of whole-genome sequence data, we examined rare and low-frequency variants for association with non-HDL cholesterol, HDL cholesterol, LDL cholesterol, and triglycerides in up to 119,146 Icelanders. We discovered 13 variants with large effects (within ANGPTL3, APOB, ABCA1, NR1H3, APOA1, LIPC, CETP, LDLR, and APOC1) and replicated 14 variants. Five variants within PCSK9, APOA 1, ANGPTL4, and LDLR associate with CAD (33,090 cases and 236,254 controls). We used genetic risk scores for the lipid fractions to examine their causal relationship with CAD. The non-HDL cholesterol genetic risk score associates most strongly with CAD (P = 2.7 × 10−28), and no other genetic risk score associates with CAD after accounting for non-HDL cholesterol. The genetic risk score for non-HDL cholesterol confers CAD risk beyond that of LDL cholesterol (P = 5.5 × 10−8), suggesting that targeting atherogenic remnant cholesterol may reduce cardiovascular risk.
Reducing LDL cholesterol levels with statins has proven effective at lowering the risk of CAD. This success has inspired the development of new therapeutics that target other lipid fractions and further reduce cardiovascular risk1-3. However, therapies that target HDL cholesterol and/or triglyceride levels have failed to consistently reduce cardiovascular events in randomized trials1-3.
Major progress in sequencing technology has enabled comprehensive detection of rare sequence variants with large effects on phenotypes. Large studies have recently yielded four such variants affecting lipid fractions4 and shown that rare variant burden within several genes5 associates with lipid traits and, in some cases, with the risk of CAD6-8. Further discoveries of such variants may promote understanding of lipid pathways and their relationship with CAD.
Sequence variants can be used as proxies for exposure to infer causality, a key factor in understanding the potential of novel therapeutics. The limitations of observational epidemiology are then minimized given the random assignment of genetic predisposition, termed Mendelian randomization. Genetic and epidemiological studies have consistently supported a direct role for LDL cholesterol in atherogenesis9-11. Recently, large studies that jointly examined the effects of sequence variants on LDL cholesterol, HDL cholesterol, and triglyceride levels concluded that triglycerides10,11, but not HDL cholesterol10-12, also have a role in the pathogenesis of CAD.
Cholesterol and triglycerides are transferred together in blood by triglyceride-rich lipoproteins, including very low-density lipoprotein (VLDL), chylomicrons, and their cholesterol-enriched remnants. A body of evidence suggests that triglycerides are not causal in the pathogenesis of CAD but are rather a marker of the atherogenic cholesterol content of triglyceride-rich lipoproteins (reviewed in ref. 13). This hypothesis can be addressed in genetic studies by accounting for effects of variants on non-HDL cholesterol (total cholesterol minus HDL cholesterol), which incorporates cholesterol from triglyceride-rich lipoproteins whereas LDL cholesterol does not.
As common sequence variants have been extensively studied in large genome-wide association studies (GWAS)11, we used a large population-based resource of whole-genome sequence data14,15 to search for new rare and low-frequency lipid-associated variants that may suggest new therapeutic targets for CAD. We subsequently used information on both rare and common variants to dissect the relationship between lipid traits and CAD.
RESULTS
Study overview
Sequence variants identified through whole-genome sequencing of 2,636 Icelanders and subsequently imputed into a large population-based data set were tested for association with non-HDL cholesterol (n = 119,146), HDL cholesterol (n = 119,514), and triglycerides (n = 80,111). For comparison, we also provide results for LDL cholesterol (n = 53,841). We focused on rare and low-frequency sequence variants (allele frequency <1% and 1–5%, respectively) annotated16 as having moderate or high impact (including missense, in-frame indel, splice-region, and loss-of-function variants) (n = 103,456). To account for prior probability17 of impact, we applied a significance threshold depending on variant class (moderate or high impact)16 of P < 1.7 × 10−7 or 3.7 × 10−6, respectively. Conditional analyses were performed to confirm that the reported rare or low-frequency variants were not explained by more strongly associated variants elsewhere in associated regions (Supplementary Table 1) and to confirm that known GWAS signals were not explained by the rare or low-frequency variants reported in our study (data not shown). The lipid-associated variants were then tested for association with CAD among 33,090 Icelandic cases and 236,254 controls.
We examined the causal relationship between lipid traits and CAD using the identified rare and low-frequency variants as well as known common and low-frequency lipid-associated variants11, specifically addressing whether the previously suggested effect of triglycerides on risk of CAD10,11 may be explained by the cholesterol content of triglyceride-rich lipoproteins.
Variant association with lipid traits
We discovered 13 rare and low-frequency variants that associated with at least one lipid trait in ANGPTL3, APOB, ABCA1, NR1H3, APOA1, LIPC, CETP, LDLR, and APOC1 (Table 1) and confirmed association of 14 previously identified variants in PCSK9, APOB, LPL, APOC3, MAP1A, CETP, LCAT, ABCA6, CD300LG, LIPG, ANGPTL4, LDLR, and HNF4A (Table 1 and Supplementary Tables 1 and 2). Many of the variants affected more than one lipid trait.
Table 1.
Association of rare and low-frequency variants with blood lipids
| Non-HDL-C (mmol/L) n = 119,146 |
HDL-C (mmol/L) n = 119,514 |
Triglycerides (% change) n = 80,111 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Variant | rsID | A1/A2 | A2 (%) | Effect (95% Cl) | P | Effect (95% Cl) | P | Effect (95% Cl) | P |
| PCSK9 | p.Arg46Leu | rs11591147 | G/T | 1.17 | −0.58 (−0.65, −0.51) |
1.2 × 10−64 | 0.031 (0.003, 0.058) |
0.028 | −1.7 (−4.7, 1.3) |
0.25 |
| ANGPTL3 a | Splice region | rs372257803 | T/C | 0.27 | −0.30 (−0.44, −0.17) |
1.4 × 10−5 | −0.053 (−0.108, 0.003) |
0.063 | −16.9 (−21.9, −11.6) |
5.7 × 10−9 |
| ANGPTL3 a | p.Asp290His | 1:62602317 | G/C | 0.49 | −0.38 (−0.48, −0.29) |
7.2 × 10−15 | −0.043 (−0.082, −0.003) |
0.033 | −20.9 (−24.4, −17.3) |
1.6 × 10−24 |
| APOB | p.His1923Arg | rs533617 | T/C | 4.52 | −0.21 (−0.24, −0.17) |
1.3 × 10−32 | 0.047 (0.033, 0.061) |
4.6 × 10−11 | −6.0 (−8.7, −3.1) |
4.7 × 10−5 |
| APOB a | p.Gly1829Glufs | 2:21011381 | TC/T | 0.007 | −3.28 (−4.24, −2.32) |
1.7 × 10−11 | 0.33 (−0.08, 0.73) |
0.12 | −65.3 (−77.7, −46.1) |
2.6 × 10−6 |
| APOB a | p.Leu1550Arg | 2:21012219 | A/C | 0.017 | −2.04 (−2.60, −1.48) |
7.8 × 10−13 | 0.34 (0.11, 0.57) |
0.004 | −35.0 (−49.1, −17.0) |
5.6 × 10−4 |
| APOB a | p.Gln725 | rs374473614 | G/A | 0.02 | −3.09 (−3.63, −2.55) |
3.2 × 10−29 | 0.31 (0.09, 0.53) |
5.9 × 10−3 | −63.2 (−71.0, −53.2) |
3.4 × 10−16 |
| LPL | p.Asp36Asn | rs1801177 | G/A | 1.22 | 0.054 (−0.010, 0.119) |
0.099 | −0.086 (−0.112, −0.060) |
1.4 × 10−10 | 9.5 (6.4, 12.7) |
7.0 × 10−10 |
| LPL | p.Asn318Ser | rs268 | A/G | 1.14 | 0.042 (−0.024, 0.109) |
0.21 | −0.10 (−0.13, −0.08) |
1.1 × 10−13 | 12.0 (8.7, 15.4) |
1.5 × 10−13 |
| ABCA1 a | Splice region | rs188308962 | A/G | 0.35 | −0.17 (−0.30, −0.05) |
6.3 × 10−3 | −0.23 (−0.28, −0.18) |
2.2 × 10−19 | −4.2 (−9.3, 1.3) |
0.13 |
| NR1H3 a | Splice region | rs200557846 | C/A | 0.61 | −0.006 (−0.096, 0.084) |
0.90 | 0.12 (0.08, 0.16) |
7.2 × 10−10 | −5.6 (−9.4, −1.7) |
5.6 × 10−3 |
| APOC3 | Splice donor | rs138326449 | G/A | 0.23 | −0.51 (−0.66, −0.36) |
2.4 × 10−11 | 0.34 (0.28, 0.40) |
1.2 × 10−27 | −44.6 (−48.3, −40.8) |
7.2 × 10−66 |
| APOA1 a | p.Val43Leu | 11:116837074 | C/G | 0.70 | −0.16 (−0.25, −0.08) |
2.5 × 10−4 | 0.17 (0.14, 0.21) |
4.5 × 10−22 | 5.2 (1.2, 9.4) |
0.01 |
| MAP1A | p.Pro2349Leu | rs55707100 | C/T | 2.76 | 0.017 (−0.028, 0.061) |
0.47 | −0.046 (−0.063, −0.028) |
3.1 × 10−7 | 6.8 (4.7, 8.9) |
3.8 × 10−11 |
| LIPC a | p.Arg208His | rs200684324 | G/A | 0.18 | 0.15 (−0.02, 0.31) |
0.079 | 0.19 (0.13, 0.26) |
1.3 × 10−8 | 14.2 (6.1, 22.8) |
4.1 × 10−4 |
| CETP a | p.Val39Glyfs | 16:56962092 | T/TG | 0.06 | −0.53 (−0.81, −0.25) |
2.4 × 10−4 | 0.40 (0.29, 0.52) |
5.4 × 10−12 | −5.0 (−16.6, 8.2) |
0.44 |
| CETP a | p.Glu443Lys | rs536221680 | G/A | 0.29 | −0.31 (−0.45, −0.18) |
6.3 × 10−6 | 0.33 (0.28, 0.39) |
4.4 × 10−32 | −0.2 (−5.6, 5.6) |
0.96 |
| CETP | p.Arg468Gln | rs1800777 | G/A | 3.57 | 0.10 (0.06, 0.13) |
1.3 × 10−6 | −0.18 (−0.19, −0.16) |
7.4 × 10−108 | 2.2 (0.5, 4.0) |
0.01 |
| LCAT | p.Ser232Thr | rs4986970 | A/T | 2.90 | 0.00 (−0.043, 0.043) |
0.99 | −0.048 (−0.066, −0.031) |
5.2 × 10−8 | 0.4 (−1.5, 2.4) |
0.67 |
| CD300LG | p.Arg82Cys | rs72836561 | C/T | 3.36 | 0.045 (0.005, 0.084) |
0.026 | −0.087 (−0.103, −0.071) |
3.2 × 10−26 | 6.7 (4.8, 8.6) |
7.3 × 10−13 |
| ABCA6 | p.Cys1359Arg | rs77542162 | A/G | 2.55 | 0.15 (0.10, 0.19) |
5.4 × 10−11 | −0.007 (−0.025, 0.010) |
0.41 | −2.6 (−4.5, −0.6) |
0.01 |
| LIPG | p.Asn396Ser | rs77960347 | A/G | 1.17 | 0.16 (0.10, 0.23) |
1.2 × 10−6 | 0.13 (0.10, 0.15) |
1.0 × 10−20 | 3.5 (0.6, 6.6) |
0.019 |
| ANGPTL4 | p.Glu40Lys | rs116843064 | G/A | 2.39 | −0.16 (−0.21, −0.11) |
8.7 × 10−11 | 0.10 (0.08, 0.12) |
3.9 × 10−23 | −11.9 (−13.7, −9.9) |
1.4 × 10−30 |
| LDLR | Splice donor | rs200238879 | T/C | 0.056 | 1.39 (1.12, 1.66) |
1.7 × 10−23 | −0.054 (−0.165, 0.057) |
0.34 | −12.0 (−22.1, −0.6) |
0.04 |
| LDLR a | Splice region | rs72658867 | G/A | 2.22 | −0.42 (−0.47, −0.37) |
5.2 × 10−63 | 0.029 (0.009, 0.049) |
4.2 × 10−3 | −1.8 (−3.9, 0.4) |
0.11 |
| APOC1 a | Intronic | rs539667984 | G/C | 0.82 | −0.12 (−0.20., −0.04) |
3.0 × 10−3 | 0.11 (0.07, 0.14) |
1.8 × 10−10 | −13.6 (−16.7, −10.4) |
4.0 × 10−15 |
| HNF4A | p.Thr139lle | rs1800961 | C/T | 4.60 | −0.04 (−0.072, −0.008) |
0.013 | −0.065 (−0.079, −0.051) |
5.2 × 10−20 | 1.0 (−0.5, 2.5) |
0.21 |
Effect (in mmol/L or percent change for log-transformed triglyceride levels) is given with respect to allele A2. Variant type was determined on the basis of information from Ensembl release 70 using Variant Effect Predictor (VEP) version 2.8. For variants without an rsID, the chromosomal position (chr:position) in Build 38 of the human reference sequence is given. Non-HDL-C, non-HDL cholesterol; HDL-C, HDL cholesterol; A1/A2, major allele/minor allele; A2 (%), frequency of allele A2; Cl, confidence interval.
New sequence variant.
Of the newly identified variants, six primarily affected HDL cholesterol. The rare allele of rs 188308962 in ABCA1 decreased HDL cholesterol levels by 0.23 mmol/L, and five rare alleles of variants in NR1H3, APOA1, LIPC, and CETP increased HDL cholesterol levels. The two CETP variants (encoding p.Val39Glyfs*62 and p.Glu443Lys) had a considerably larger effect on HDL cholesterol levels (increases of 0.40 and 0.33 mmol/L, respectively) than the NR1H3, APOA1, and LIPC variants (increases of 0.12, 0.17, and 0.19 mmol/L, respectively) (Table 1).
Four new variants mainly affected non-HDL cholesterol. A low-frequency splice-region allele at rs72658867 within the LDL receptor gene LDLR lowered non-HDL cholesterol levels by 0.42 mmol/L, and three extremely rare mutations (frequency of 0.01–0.02%) in the apolipoprotein B gene APOB (encoding p.Giy1829Giufs*8, p.Leu 1550Arg, and p.Gln725*) each lowered non-HDL cholesterol levels markedly (by 3.28, 2.04, and 3.09 mmol/L, respectively). All phenotyped carriers of these APOB mutations had extremely low non-HDL cholesterol and triglyceride levels, particularly carriers of the p.Gly1829Glufs*8 and p.Gln725* variants, indicating that these represent new familial mutations associated with hypobetalipoproteinemia (Supplementary Fig. 1). The burden of loss-of-function mutations in APOB, including the p.Gln725* variant seen in a single individual, was recently shown to reduce levels of LDL cholesterol5.
Two variants within ANGPTL3 (rs372257803 and p.Asp290His) mainly affected triglycerides, with the rare allele decreasing levels by 16.9% and 20.9% (corresponding to decreasing the level of an individual with the population mean by 0.22 and 0.27 mmol/L, respectively), but these variants also lowered non-HDL cholesterol levels by 0.30 and 0.38 mmol/L.
Finally, we observed an association of a missense variant in ZNF285 (encoding p.Gly480Arg) with triglycerides and HDL cholesterol. However, conditional analyses showed that the ZNF285 variant is explained by a rare intronic variant (rs539667984) in APOC1 that lowered triglyceride levels by 13.6% (corresponding to decreasing the level of an individual with the population mean by 0.17 mmol/L) and increased HDL cholesterol levels by 0.11 mmol/L (Table 1 and Supplementary Tables 1 and 2). We replicated the recently reported4 association between rs186808413 in PAFAH1B2 and both HDL cholesterol and triglycerides. However, we show that this association is largely explained by the APOC3 splice donor variant6,7 rs138326449 (Supplementary Table 1).
For follow-up in populations from the Netherlands (n = 5,473) and Iran (n = 9,491), we prioritized the genotyping of variants that were likely to be polymorphic outside Iceland (reported in dbSNP18 or the Exome Aggregation Consortium (ExAC)). We observed association with the same lipid traits as in Iceland for the new rs188308962 (ABCA1) and rs72658867 (LDLR) variants and for the previously reported rs138326449 (APOC3) variant (Supplementary Table 3). The remaining variants were extremely rare in the outside populations, resulting in insufficient power to detect association.
The 185 variants previously reported to associate with various lipid traits11 account for 7.7%, 9.8%, and 5.7% of the population variance in Iceland in the levels of non-HDL cholesterol, HDL cholesterol, and triglycerides, respectively, and the 27 rare and low-frequency variants reported in our study account for an additional 4.1%, 4.1%, and 2.4%, respectively.
Variant association with coronary artery disease
Of the 27 rare and low-frequency lipid-associated variants identified in our study, 5 associated with CAD in Iceland (Table 2 and Supplementary Table 2) when adjusting for 27 tests (P < 1.7 × 10−3). Conditioning on known signals from lipid GWAS in the respective loci had negligible effects on the association of the five variants with CAD (data not shown). Carriers of the LDLR splice donor mutation at rs200238879 had 2.8-fold higher risk of CAD, whereas the minor allele of LDLR splice-region variant rs72658867 and the variant in PCSK9 encoding p.Arg46Leu decreased risk by 23% and 27%, respectively. The APOA1 variant encoding p.Val43Leu that primarily increased HDL cholesterol levels and the ANGPTL4 variant encoding p.Glu40Lys that primarily lowered triglyceride and increased HDL cholesterol levels decreased risk of CAD by 26% and 20%, respectively. The APOA1 and ANGPTL4 variants also affected non-HDL cholesterol levels to a lesser degree (each resulting in a decrease of0.16 mmol/L). All five variants affected age at diagnosis in a direction consistent with their effect on CAD (Table 2).
Table 2.
Effect of variants that associate with coronary artery disease on age at diagnosis and lifespan
| CAD (n = 33,090 cases, 236,254 controls) |
Age at diagnosis of CAD (n = 33,090) |
Lifespan (n = 61,060) |
|||||
|---|---|---|---|---|---|---|---|
| Variant | A2 (%) | OR | P | β | P | β | P |
| PCSK9 (rs11591147) | 1.17 | 0.73 | 3.1 × 10−6 | 2.44 | 0.0005 | 0.78 | 0.081 |
| APOA1 (p.Val43Leu) | 0.70 | 0.74 | 0.0004 | 2.00 | 0.027 | 1.40 | 0.015 |
| ANGPTL4 (rs116843064) | 2.39 | 0.80 | 3.4 × 10−6 | 0.97 | 0.047 | 0.28 | 0.38 |
| LDLR (rs200238879) | 0.056 | 2.76 | 0.0003 | −8.83 | 0.0001 | −6.54 | 0.0010 |
| LDLR (rs72658867) | 2.22 | 0.77 | 1.0 × 10−7 | 1.18 | 0.018 | 0.54 | 0.09 |
All effects (odds ratio and β) are given with respect to allele A2 (allele information appears in Table 1). Effects (β) for association with age at diagnosis of CAD and lifespan are in years. Lifespan (number of years lived) is the mean age at death of individuals born after 1890 who lived to be at least 50 years old. The APOA1 variant encoding p.Val43Leu is located on chromosome 11 at 116,837,074 bp in Build 38 of the human reference sequence. CAD, coronary artery disease; OR, odds ratio; A2 (%), frequency of allele A2.
An aggregate of loss-of-function mutations in APOC3, primarily driven by rs138326449, was recently reported to associate with CAD6,7. There was no association between rs138326449 and CAD in Iceland (Supplementary Tables 2 and 4). Although our results do not support the reported effects of APOC3 loss-of-function mutations on risk of CAD, a more modest effect cannot be excluded.
Blood lipids causing coronary artery disease
We constructed genetic risk scores for each lipid trait based on the 27 rare and low-frequency lipid-associated variants identified in this study and the 185 reported common and low-frequency lipid-associated variants11. Genetic risk scores for non-HDL cholesterol, HDL cholesterol, LDL cholesterol, and triglycerides all associated individually with CAD (Table 3). As expected, the CAD risk associated with the LDL cholesterol genetic risk score was fully captured by that of non-HDL cholesterol (P = 0.075; Table 3). In contrast, the genetic risk score for non-HDL cholesterol conferred additional risk of CAD after accounting for the risk associated with the LDL cholesterol genetic risk score (P = 5.5 × 10−8) (Table 3).
Table 3.
Association of genetic risk scores with coronary artery disease
| GRS 1 |
GRS 2 |
GRS-all |
||||||
|---|---|---|---|---|---|---|---|---|
| Outcome | Predictor | Covariate | β | P | β | P | β | P |
| CAD | Non-HDL-C GRS | – | 0.44 | 7.6 × 10−14 | 0.27 | 4.6 × 10−19 | 0.28 | 2.7 × 10−28 |
| HDL-C GRS, TG GRS | 0.45 | 5.8 × 10−11 | 0.23 | 2.4 × 10−11 | 0.26 | 4.4 × 10−18 | ||
| LDL-C GRS | 0.56 | 3.2 × 10−4 | 0.36 | 3.8 × 10−5 | 0.41 | 5.5 × 10−8 | ||
| CAD | LDL GRS | – | 0.40 | 2.8 × 10−11 | 0.24 | 1.3 × 10−15 | 0.25 | 1.4 × 10−22 |
| HDL-C GRS, TG GRS | 0.36 | 6.7 × 10−9 | 0.21 | 1.6 × 10−11 | 0.22 | 7.4 × 10−17 | ||
| Non-HDL-C GRS | −0.13 | 0.40 | −0.10 | 0.25 | −0.13 | 0.075 | ||
| CAD | HDL-C GRS | – | −0.17 | 0.0001 | −0.09 | 0.0006 | −0.09 | 9.0 × 10−6 |
| Non-HDL-C GRS, TG GRS | −0.08 | 0.13 | −0.016 | 0.57 | −0.017 | 0.44 | ||
| LDL-C GRS, TG GRS | −0.09 | 0.08 | −0.020 | 0.48 | −0.022 | 0.34 | ||
| CAD | TG GRS | – | 0.22 | 0.0007 | 0.21 | 6.4 × 10−10 | 0.20 | 4.9 × 10−12 |
| Non-HDL-C GRS, HDL-C GRS | −0.10 | 0.19 | 0.07 | 0.11 | 0.042 | 0.24 | ||
| LDL-C GRS, HDL-C GRS | 0.06 | 0.43 | 0.14 | 0.0002 | 0.13 | 7.5 × 10−5 | ||
GRSs were calculated using data from 104,131 individuals (15,575 CAD cases and 88,556 controls). GRS 1 was based on 27 rare and low-frequency lipid-associated variants identified in this study. GRS 2 was based on 185 known lipid-associated variants. GRS-all is the combination of GRS 1 and GRS 2. The effect (β) for CAD is the logarithm of the odds ratio for CAD per 1 s.d. of the predictor. CAD, coronary artery disease; GRS, genetic risk score; non-HDL-C, non-HDL cholesterol; LDL-C, LDL cholesterol; HDL-C, HDL cholesterol; TG, triglycerides.
In joint analysis of non-HDL cholesterol, HDL cholesterol, and triglycerides, after adjusting for the genetic risk scores of the other lipid traits, only the association of non-HDL cholesterol with CAD remained significant (P value for the genetic risk score including all variants (Pall) = 4.4 × 10−18, 0.44, and 0.24 for non-HDL cholesterol, HDL cholesterol, and triglycerides, respectively). Further, the genetic risk score for non-HDL cholesterol was associated with earlier age at diagnosis of CAD (2.55 years per unit of the genetic risk score. Pall = 3.9 × 10−13) and shorter lifespan (1.63 years per unit of the genetic risk score, Pall = 6.0 × 10−18) (Table 4).
Table 4.
Association of genetic risk scores with age at diagnosis of CAD and lifespan
| GRS 1 |
GRS 2 |
GRS-all |
||||||
|---|---|---|---|---|---|---|---|---|
| Outcome | Predictor | Covariate | β | P | β | P | β | P |
| Age at diagnosis of CAD | Non-HDL-C GRS | – | −3.92 | 7.8 × 10−9 | −2.58 | 4.3 × 10−13 | −2.7 | 9.8 × 10−19 |
| HDL-C GRS, TG GRS | −3.57 | 8.5 × 10−6 | −2.49 | 1.3 × 10−9 | −2.55 | 3.9 × 10−13 | ||
| LDL-C GRS | −6.07 | 9.8 × 10−4 | −1.63 | 0.121 | −2.80 | 0.002 | ||
| LDL GRS | – | −3.45 | 9.3 × 10−7 | −2.52 | 9.1 × 10−13 | −2.516 | 1.2 × 10−16 | |
| HDL-C GRS, TG GRS | −2.91 | 6.6 × 10−5 | −2.32 | 1.3 × 10−10 | −2.278 | 2.6 × 10−13 | ||
| Non-HDL-C GRS | 2.41 | 0.21 | −1.00 | 0.33 | 0.080 | 0.93 | ||
| HDL-C GRS | – | 2.00 | 2.7 × 10−4 | 0.79 | 0.0113 | 0.893 | 2.8 × 10−4 | |
| Non-HDL-C GRS, TG GRS | 1.05 | 0.096 | 0.25 | 0.47 | 0.28 | 0.30 | ||
| LDL-C GRS, TG GRS | 1.16 | 0.066 | 0.29 | 0.40 | 0.32 | 0.24 | ||
| TG GRS | – | −2.45 | 0.001 | −1.57 | 9.2 × 10−5 | −1.71 | 8.9 × 10−7 | |
| Non-HDL-C GRS, HDL GRS | 0.26 | 0.78 | −0.04 | 0.93 | −0.12 | 0.78 | ||
| LDL-C GRS, HDL GRS | −0.99 | 0.250 | −0.84 | 0.062 | −0.96 | 0.014 | ||
| Lifespan | Non-HDL-C GRS | – | −2.45 | 8.9 × 10−6 | −1.28 | 1.6 × 10−5 | −1.52 | 8.3 × 10−9 |
| HDL-C GRS, TG GRS | −2.22 | 0.0008 | −1.52 | 9.2 × 10−6 | −1.63 | 6.0 × 10−8 | ||
| LDL-C GRS | −2.44 | 0.11 | −0.047 | 0.96 | −0.755 | 0.32 | ||
| LDL GRS | – | −2.35 | 3.4 × 10−5 | −1.36 | 5.1 × 10−6 | −1.46 | 8.1 × 10−9 | |
| HDL-C GRS, TG GRS | −2.02 | 7.4 × 10−4 | −1.37 | 7.9 × 10−6 | −1.43 | 5.7 × 10−8 | ||
| Non-HDL-C GRS | 0.02 | 0.99 | −1.32 | 0.14 | −0.77 | 0.32 | ||
| HDL-C GRS | – | 0.93 | 0.04 | −0.058 | 0.81 | 0.15 | 0.46 | |
| Non-HDL-C GRS, TG GRS | 0.20 | 0.70 | −0.27 | 0.36 | −0.16 | 0.49 | ||
| LDL-C GRS, TG GRS | 0.25 | 0.64 | −0.24 | 0.41 | −0.13 | 0.56 | ||
| TG GRS | – | −1.75 | 0.005 | −0.34 | 0.32 | −0.64 | 0.027 | |
| Non-HDL-C GRS, HDL GRS | −0.16 | 0.84 | 0.39 | 0.36 | 0.18 | 0.62 | ||
| LDL-C GRS, HDL GRS | −0.93 | 0.20 | −0.12 | 0.75 | −0.35 | 0.29 | ||
GRSs were calculated using data from 15,575 individuals with information on age at diagnosis of CAD; information on lifespan was available for 16,305 individuals. GRS 1 was based on 27 rare and low-frequency lipid-associated variants identified in this study. GRS 2 was based on 185 known lipid-associated variants. GRS-all was the combination of GRS 1 and GRS 2. The effects (β) for age at diagnosis of CAD (adjusted for sex) and lifespan are in years. GRS, genetic risk score; non-HDL-C, non-HDL cholesterol; LDL-C, LDL cholesterol; HDL-C, HDL cholesterol; TG, triglycerides.
Previous Mendelian randomization studies10,11 that concluded that triglycerides have a causal role in CAD used LDL rather than non-HDL cholesterol in their analysis, ignoring the atherogenic cholesterol contained by triglyceride-rich lipoproteins. We replicated a residual association between triglycerides and CAD when LDL cholesterol was used in the causal analysis, but this association was no longer seen after also accounting for remnant cholesterol through non-HDL cholesterol (Table 3).
To compare our results using individual-level genetic risk scores with results based on summary-level data for Mendelian randomization, we applied two variations of weighted multiple regression: a modified approach used by Do et al.11 and an extension to multiple exposures of the framework proposed by Bowden et al.19 in which the effect of each variant is based on the allele that has a positive effect on the blood lipid level. All methods showed that the CAD risk associated with these lipid traits was best and fully captured by non-HDL cholesterol, with no independent association with HDL cholesterol or triglycerides (Supplementary Tables 5 and 6). Further, all methods agreed that non-HDL cholesterol confers additional risk of CAD beyond that from LDL cholesterol (Supplementary Tables 5 and 6).
Reanalysis of the data of Do et al., where a causal link between triglycerides and CAD was claimed, accounting for non-HDL instead of LDL cholesterol levels using weighted multiple linear regression did not support an independent effect for triglycerides on CAD risk (Supplementary Note).
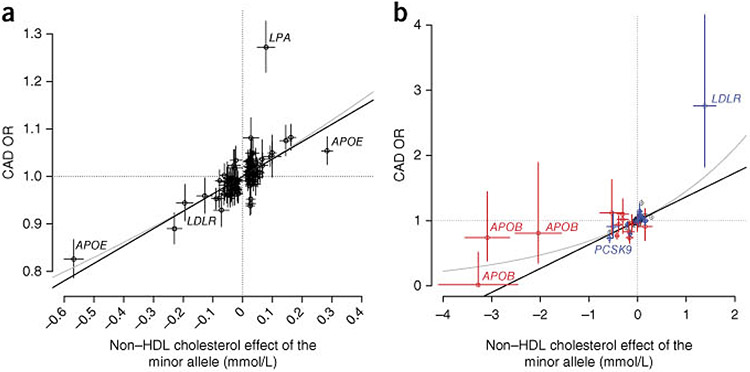
We plotted the relationship between variant effects on non-HDL cholesterol levels and the associated effects on CAD (Fig. 1). Although there was a strong positive correlation between levels of non-HDL cholesterol and degree of CAD risk, disparities in effect magnitudes were frequently observed.
Figure 1.
Relationship between the effect of sequence variants on non-HDL cholesterol and their effect on risk of coronary artery disease, (a) Effect on CAD risk of 100 known common (minor allele frequency (MAF) >5%) lipid-associated variants11,29,30 that associate with non-HDL cholesterol in Iceland (P < 0.05). (b) Effect on CAD of the 27 rare and low-frequency lipid-associated variants identified in this study. Previously published variants are colored blue, and new variants are colored red. In both plots, the effect on non-HDL cholesterol (n = 119,146) is given in mmol/L, and the effect on CAD risk (n = 33,090 cases and 236,254 controls) is represented by the odds ratio (OR). Bars represent 95% confidence intervals for the effect on non-HDL cholesterol and risk of CAD. The black line is the line best fitting the common variant, and the gray line is the line best fitting the CAD odds ratio on the log scale transformed back to the original scale.
DISCUSSION
We discovered 13 rare and low-frequency sequence variants with large effects on lipid levels and confirmed 14 previously reported ones. Some of these variants are mutations with large effects, presumably causing Mendelian dyslipidemia, including the splice donor mutation (rs200238879) in LDLR, which was previously described as causing familial hypercholesterolemia20 in Iceland. We also discovered two highly penetrant APOB mutations, encoding p.Gln725* and p.Gly1829Glufs*8, causing familial hypobetalipoproteinemia.
The newly identified variants all reside within genes with strong evidence for roles in lipid metabolism (Supplementary Fig. 2), including extensive literature support from analysis of familial mutations within respective genes, characterization of transgenic mice, and biochemical studies9,21-24. Many of the new variants have similar effects on the lipid profile as known loss-of-function mutations within these genes, such as the CETP mutations21 encoding p.Val39Glyfs*62 and p.Glu443Lys. In contrast, whereas over 40 rare APOA1 mutations that lower HDL cholesterol levels are known21, rare mutations of APOA1 that increase HDL cholesterol levels, such as the p.Val43Leu variant, to our knowledge have not been reported before.
The results of our causality analysis applying genetic risk scores based on the 27 rare and low-frequency variants reported in this study combined with 185 known lipid-associated variants11 were consistent with an effect of non-HDL cholesterol, but not of HDL cholesterol or triglycerides, on the pathogenesis of CAD. This is in agreement with epidemiologic data from the Emerging Risk Factors Collaboration, based on 2.79 million person–years of follow-up of 302,430 individuals, showing no association between triglycerides and CAD after adjustments for HDL cholesterol and non-HDL cholesterol25. Our findings are also consistent with a recent genetic study that supports a causative role for remnant cholesterol (defined as non-HDL cholesterol minus LDL cholesterol) in CAD13. Consistent with our reanalysis of the data of Do et al. (Supplementary Note), we show that the observed association between triglycerides and CAD can be explained by the effect of cholesterol carried within triglyceride-rich lipoproteins, captured by non-HDL cholesterol, and that non-HDL cholesterol confers additional risk of CAD over the risk associated with LDL cholesterol. Therefore, targeting atherogenic cholesterol beyond that carried by LDL particles may represent a promising strategy for further reduction of cardiovascular risk.
Although there is a strong correlation between the effects of sequence variants on non-HDL cholesterol and risk of CAD, disparities in effect magnitude are evident, which can probably be explained by a variety of pleiotropic effects26 that extend beyond the crude lipid measures used in our study and in clinical practice. More detailed phenotyping, such as lipoprotein subfraction profiling, may provide more information on the causal role of lipid traits in cardiovascular disease. Furthermore, although general causal relationships between HDL cholesterol or triglyceride levels and CAD are not supported, the possibility remains that certain mechanisms that affect these variables could have a role in the pathogenesis of the disease. Indeed, we do find that two variants that primarily affect HDL cholesterol and/or triglycerides, the APOA1 variant encoding p.Val43Leu and the ANGPTL4 variant encoding p.Glu40Lys, have considerable effects on CAD, pointing to the products of these genes as potential therapeutic targets. Their effects on CAD may be explained by their impact on non-HDL cholesterol, although other mechanisms27 could have a role.
In conclusion, we have discovered 13 rare and low-frequency sequence variants with large effects on lipid levels and replicated 14 other such variants. The identification of rare variants underlying common traits is facilitated by the reduced allele diversity in Iceland28 relative to larger and less homogenous populations. Our results are consistent with a role of non-HDL cholesterol, but not of HDL cholesterol or triglycerides, in the pathogenesis of CAD.
ONLINE METHODS
Study governance.
The Icelandic study and the studies from the Netherlands, Iran, and the United States were approved by the appropriate bioethics and/or data protection authorities. All participating subjects donating blood signed informed consent forms. The personal identities of the individuals from whom phenotype information and biological samples were obtained were encrypted by a third-party system provided by the Icelandic Data Protection Authority.
Participants.
Iceland.
Participants (Supplementary Table 7) were enrolled as part of various genetics programs at deCODE. Lipid measurements were obtained from three of the largest clinical laboratories in Iceland: (i) Landspítali–National University Hospital (LUH), Reykjavík (hospitalized and ambulatory patients); (ii) the Laboratory in Mjódd (RAM), Reykjavík (ambulatory patients); and (iii) Akureyri Hospital, Regional Hospital in North Iceland, Akureyri (hospitalized and ambulatory patients). LDL cholesterol concentration was calculated using the Friedewald equation31 (for triglyceride levels <4.00 mmol/L), and non-HDL cholesterol concentration was calculated by subtracting HDL cholesterol levels from total cholesterol levels. CAD cases were defined as (i) individuals in the MONICA registry32 who suffered myocardial infarction before the age of 75 years in Iceland between 1981 and 2002; (ii) subjects with discharge diagnoses of CAD (ICD-9 codes 410.*, 411.*, 412.*, and 414.* or ICD-10 codes I20.0, I21.*, I22.*, I23.*, I24.*, and I25.*) from LUH; (iii) subjects diagnosed with significant angiographic CAD (at least 50% luminal reduction33) identified from a nationwide clinical registry of coronary angiography and percutaneous coronary interventions at LUH between 1987 and 2012; (iv) subjects undergoing coronary artery bypass grafting (CABG) procedures at LUH between 2002 and 2011; or (v) individuals with cause of death or a contributing cause of death listed as myocardial infarction or CAD on death registries between 1996 and 2009. Coronary angiograms were evaluated by interventional cardiologists. The lifespan variable was based on individuals who were born after 1890 and lived to be at least 50 years old.
The Netherlands.
Subjects from the Netherlands were recruited within the ‘Nijmegen Biomedical Study’, a population-based survey in which age- and sex-stratified adult residents of Nijmegen were invited to participate34.
Iran.
The Iranian subjects are part of the ongoing Tehran Lipid and Glucose Study35 including 15,005 residents of Tehran. For the current study, individuals ≥18 years old were included. CAD events and diagnoses (ICD-10 codes I20.0, I21.*, I22.*, I23.*, I24.*, and I25.*) were confirmed by reviewing hospital records, death certificates, or autopsy reports36.
Atlanta, USA.
Participants were enrolled through the Emory Cardiovascular Biobank Study and Clinical Registry in Neurology37. CAD was defined as significant luminal stenosis (≥50%) on coronary angiography33. Additional patients (8%) who did not meet criteria for significant angiographic CAD but had self-reported history of myocardial infarction, CABG, or percutaneous coronary intervention were included as CAD cases. Controls had no or minimal (<20%) stenosis on angiography and no history of CAD. Additional controls included individuals with non-vascular neurological diseases (mainly Parkinson or Alzheimer disease), unrelated friends of cases, and community volunteers, excluding those with a known history of CAD. The study was restricted to self-reported Caucasians.
Data handling.
Lipid levels were adjusted for sex, year of birth and age at measurement, lipid-lowering medication, and measurement site, using the average of multiple measurements for an individual, and were then normalized to a standard normal distribution using quantile normalization. To obtain effect estimates in mmol/L, the estimates from the regression analysis were multiplied by the estimated standard deviation in lipid levels in the population. Given an approximately log-normal distribution, triglyceride levels were log transformed before adjustment, and the corresponding effect estimates are presented as percentage change instead of in units of mmol/L.
Genetic risk scores.
Individual-level genetic risk scores were constructed for each lipid trait by combining the minor allele counts for the lipid-associated variants weighted by the estimated effect of each allele on the lipid trait in the Icelandic population. We imputed missing genotypes on the basis of the expected allele count. Three sets of risk scores were generated based on (i) the 27 rare and low-frequency lipid-associated variants identified in this study (GRS 1); (ii) 185 known lipid-associated variants (GRS 2)11; and (iii) the combination of GRS 1 and GRS 2 (GRS-all). To examine the impact of concomitant statin therapy on the association of genetic risk scores with CAD, we further constructed modified genetic risk scores (GRS-modified) in which each variant was weighted by its effect on the lipid trait in individuals not using statins. These modified GRSs were then tested for association with CAD (Supplementary Table 8).
Summary-level Mendelian randomization.
We examined variant effect estimates for each lipid trait on the effect estimate for CAD in our data with weighted multiple regression using 1/(SE)2 as weights, where SE is the standard error. The summary-level data were analyzed both in an approach modified from Do et al.11 and an extension to multiple exposures of the framework proposed by Bowden et al.19 in which the effect of each variant is based on the allele with a positive effect on the blood lipid level (Supplementary Tables 5 and 6, and Supplementary Note).
Fraction of variance explained.
The fraction of the variance in a quantitative trait that is explained in a population by a variant with MAF f and an effect β measured in standard units is 2f(1 – f)β2.
Genotyping.
Illumina BeadChip genotyping of Icelandic samples was performed on a HumanHap300, HumanCNV370, HumanHap610, HumanHap1M,, HumanHap660, Omnil, Omni2.5, or OmniExpress BeadChip at deCODE Genetics15. Single-track genotyping using the Centaurus (Nanogen) platform38 or Sanger sequencing was performed to validate and improve imputation of reported variants in Iceland and for association testing in other populations. Whole-genome sequencing was performed for 2,636 individuals selected for various conditions. All individuals were sequenced to a minimum depth of 10× (average of 22×)15. Paired-end libraries for sequencing were prepared according to the manufacturer’s instructions (Illumina, TruSeq). Sequencing by synthesis was performed on Illumina Genome Analyzer IIx and/or HiSeq 2000 instruments, and reads were aligned to NCBI Build 36 of the human reference sequence using Burrows–Wheeler Aligner (BWA) 0.5.9 (ref. 39).
Long-range phasing and genotype imputation.
Sequence variants identified through whole-genome sequencing were imputed into 104,220 chip-genotyped and long-range-phased Icelanders and, with the aid of Icelandic genealogical information, into 294,212 first- and second-degree relatives of array-genotyped individuals14,15,40. To improve the imputation quality of rare mutations (n = 33,026) with potential effect on lipids (those carried by 135 Icelandic exome-sequenced individuals with extreme lipid levels (below the 1st or above the 99th percentile)), the imputation process was repeated after adding genotypes from 728 exome-scqucnccd individuals and/or genotypes from 90–400 individuals obtained through single-track genotyping or Sanger sequencing to the imputation training set.
Association analysis.
A generalized form of linear or logistic regression that accounts for relatedness between individuals and potential population stratification was used to test for the association of quantitative and binary traits, respectively, with sequence variants41. Other available characteristics that correlate with disease status were also included in the model as nuisance variables. These characteristics were sex, county of birth, current age or age at death (first- and second-order terms included), blood sample availability for the individual, and an indicator function for the overlap of the lifetime of the individual with the timespan of phenotype collection. Conditional analyses were performed by including variants or genetic risk scores as covariates in the model. Association of genetic variants with lipid traits and CAD in replication samples was tested using R software.
Thresholds for genome-wide significance and genomic control correction factors.
We applied thresholds for genome-wide significance that depended on the variant class to account for prior importance of sequence variants, as described previously17. We performed a weighted Holm–Bonferroni correction based on giving equal weight to the classes annotated16 as high-impact or loss-of-function variants (stop gain, frameshift indel, and splice site), moderate-impact variants (missense and in-frame indel), and other variants. Whole-genome sequencing of 2,636 Icelanders identified 4,499 and 98,957 high- and moderate-impact variants, respectively, that passed our quality thresholds. The sum of the weights over all the variants in the genome is 1, and the Bonferroni threshold for significance within a class containing m sequence variants will be 0.05/3m. We thus used significance thresholds of P < 3.7 × 10−6 and 1.7 × 10−7 for high- and moderate-impact variants, respectively. The genomic control correction factors for non-HDL cholesterol, HDL cholesterol, triglycerides, CAD, age at diagnosis of CAD, and lifespan were 1.36, 1.40, 1.58, 1.71, 1.41, and 1.49, respectively.
Gene and variant annotation.
Sequence variants were annotated with information from Ensembl release 70 using Variant Effect Predictor (VEP) version 2.8 (ref. 16).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank all the individuals who participated in this study and whose contribution made this work possible. We also thank our valued colleagues who contributed to the data collection and phenotypic characterization of clinical samples as well as to the genotyping and analysis of the whole-genome association data. Funding was provided by deCODE Genetics/Amgen and at Emory by NIH grants UL1RR025008 from the Clinical and Translational Science Award program, R01HL089650-02, and Emory Neuroscience NINDS Core Facilities grant P30NS055077.
Footnotes
URLs. Exome Aggregation Consortium (ExAC), http://exac.broadinstitute.org/.
METHODS
Methods and any associated references are available in the online version of the paper.
Accession codes. All significant genome-wide findings are presented in Supplementary Tables 1 and 2.
Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Barter PJ et al. Effects of torcetrapib in patients at high risk for coronary events. N. Engl. J. Med 357, 2109–2122 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Schwartz GG et al. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N. Engl. J. Med 367, 2089–2099 (2012). [DOI] [PubMed] [Google Scholar]
- 3.HPS2-THRIVE Collaborative Group. Effects of extended-release niacin with laropiprant in high-risk patients. N. Engl. J. Med 371, 203–212 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Peloso GM et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am. J. Hum. Genet 94, 223–232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lange LA et al. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am. J. Hum. Genet 94, 233–245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG & Tybjærg-Hansen A Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N. Engl. J. Med 371, 32–41 (2014). [DOI] [PubMed] [Google Scholar]
- 7.TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N. Engl. J. Med 371, 22–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Do R et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature 518, 102–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown MS & Goldstein JL A receptor-mediated pathway for cholesterol homeostasis. Science 232, 34–47 (1986). [DOI] [PubMed] [Google Scholar]
- 10.Holmes MV et al. Mendelian randomization of blood lipids for coronary heart disease. Eur. Heart J 36, 539–550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Do R et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat. Genet 45, 1345–1352 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voight BF et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet 380, 572–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varbo A et al. Remnant cholesterol as a causal risk factor for ischemic heart disease. J. Am. Coll. Cardiol 61, 427–436 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Kong A et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat. Genet 40, 1068–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Styrkarsdottir U et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature 497, 517–520 (2013). [DOI] [PubMed] [Google Scholar]
- 16.McLaren W et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26, 2069–2070 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sveinbjornsson G et al. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat. Genet 48, 314–317 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Sherry ST et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowden J, Davey Smith G & Burgess S Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol 44, 512–525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudnason V, Sigurdsson G, Nissen H & Humphries SE Common founder mutation in the LDL receptor gene causing familial hypercholesterolemia in the Icelandic population. Hum. Mutat 10, 36–44 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Brunham LR & Hayden MR Human genetics of HDL: insight into particle metabolism and function. Prog. Lipid Res 58, 14–25 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Whitfield AJ, Barrett PHR, van Bockxmeer FM & Burnett JR Lipid disorders and mutations in the APOB gene. Clin. Chem 50, 1725–1732 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Berbée JFP, van der Hoogt CC, Sundararaman D, Havekes LM & Rensen PCN Severe hypertriglyceridemia in human APOC1 transgenic mice is caused by apoC-l–induced inhibition of LPL. J. Lipid Res 46, 297–306 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Tontonoz P & Mangelsdorf DJ Liver X receptor signaling pathways in cardiovascular disease. Mol. Endocrinol 17, 985–993 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Emerging Risk Factors Collaboration. Major lipids, apolipoproteins, and risk of vascular disease. J. Am. Med. Assoc 302, 1993–2000 (2009). [Google Scholar]
- 26.Würtz P et al. Lipoprotein subclass pleiotropy reveals pleiotropy in the genetic variants of lipid risk factors for coronary heart disease: a note on Mendelian randomization studies. J. Am. Coll. Cardiol 62, 1906–1908 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Rohatgi A et al. HDL cholesterol efflux capacity and incident cardiovascular events. N. Engl. J. Med 371, 2383–2393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helgason A, Nicholson G, Stefánsson K & Donnelly P A reassessment of genetic diversity in Icelanders: strong evidence from multiple loci for relative homogeneity caused by genetic drift. Ann. Hum. Genet 67, 281–297 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Clarke R et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med 361, 2518–2528 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Bennet AM et al. Association of apolipoprotein E genotypes with lipid levels and coronary risk. J. Am. Med. Assoc 298, 1300–1311 (2007). [Google Scholar]
- 31.Friedewald WT, Levy RI & Fredrickson DS Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem 18, 499–502 (1972). [PubMed] [Google Scholar]
- 32.Joint International Society and Federation of Cardiology/World Health Organization Task Force on Standardization of Clinical Nomenclature. Nomenclature and criteria for diagnosis of ischemic heart disease. Circulation 59, 607–609 (1979). [DOI] [PubMed] [Google Scholar]
- 33.Scanlon PJ et al. ACC/AHA guidelines for coronary angiography: executive summary and recommendations. A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Committee on Coronary Angiography) developed in collaboration. Circulation 99, 2345–2357 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Wetzels JFM, Kiemeney LA, Swinkels DW, Willems HL & den Heijer M Age- and gender-specific reference values of estimated GFR in Caucasians: the Nijmegen Biomedical Study. Kidney Int. 72, 632–637 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Azizi F et al. Prevention of non-communicable disease in a population in nutrition transition: Tehran Lipid and Glucose Study phase II. Trials 10, 5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khalili D et al. The incidence of coronary heart disease and the population attributable fraction of its risk factors in Tehran: a 10-year population-based cohort study. PLoS One 9, e105804 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helgadottir A et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316, 1491–1493 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Kutyavin IV et al. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res. 34, e128 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H & Durbin R Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong A et al. Parental origin of sequence variants associated with complex diseases. Nature 462, 868–874 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinthorsdottir V et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat. Genet 46, 294–298 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.