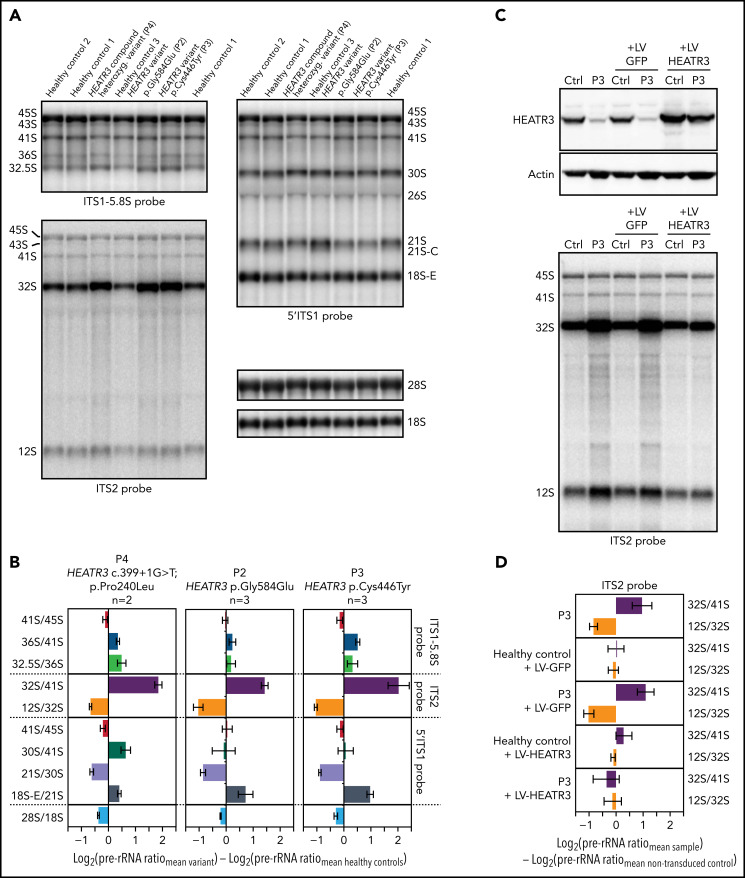
Figure 4.
Variants in HEATR3 affect pre-rRNA processing. (A) Northern blot analysis of LCLs derived from control or affected individuals carrying HEATR3 variants. Radiolabeled probes targeting the 5′ extremity of internal transcribed spacer 1 (5′ITS1), the ITS1/5.8S junction (ITS1-5.8S), or internal transcribed spacer 2 (ITS2) were used to detect pre-rRNA precursors. Each lane was loaded with 3 µg total RNA. (B) Quantification of pre-rRNAs in LCLs derived from individuals carrying HEATR3 variants was performed by using Ratio Analysis of Multiple Precursors: product-to-precursor ratios at various processing steps are expressed as variations relative to LCLs from healthy controls. Mean values ± standard error of the mean from 3 independent experiments, aside from sample P4, for which n = 2. (C) Restoration of HEATR3 expression in LCLs from P3. LCLs from P3 and from a control individual were transduced with lentiviruses expressing either green fluorescent protein (LV-GFP) or HEATR3 (LV-HEATR3). Western blot analysis confirmed expression of HEATR3 in P3 LCLs transduced with LV-HEATR3 (upper panel). Northern blot analysis of total pre-rRNAs with probe ITS2 showed restoration of a normal pre-rRNA processing pattern. (D) Ratio Analysis of Multiple Precursors quantification of the northern blot shown in panel C. Product-to-precursor ratios in P3 LCLs at steps related to the processing of the large subunit rRNA precursors are expressed as variations relative to LCLs from the healthy control. Mean values ± standard error of the mean from 3 independent experiments.