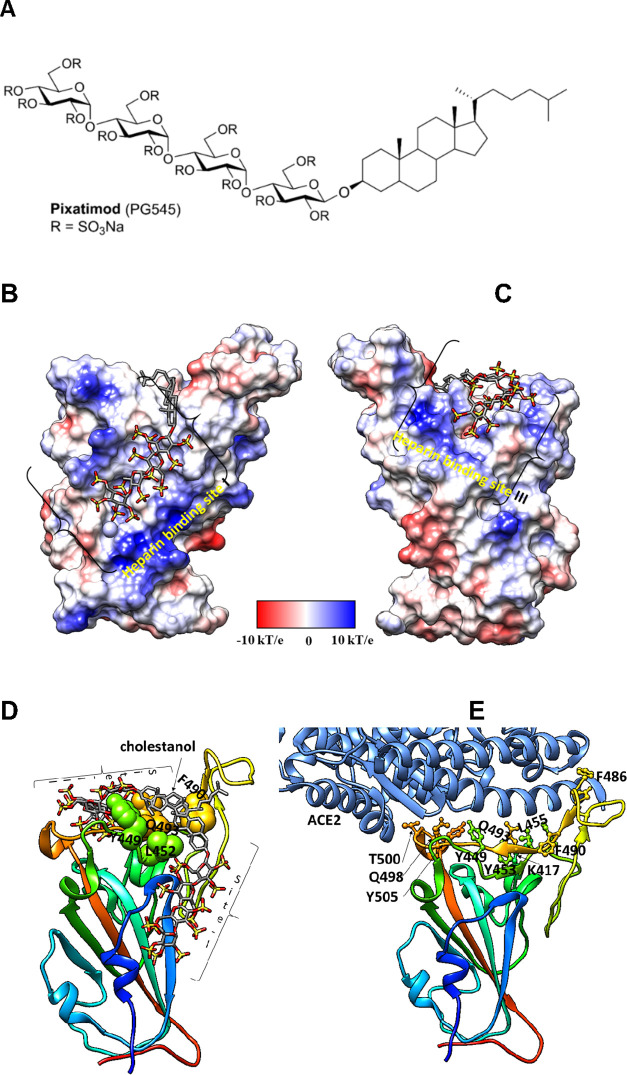
Figure 1.
Molecular dynamics modeling defines direct interactions of pixatimod with S1 RBD: (A) Structure of pixatimod. (B) Model (pose-a) showing interactions of pixatimod on the Coulombic surface of the RBD domain of spike protein. The sulfated tetrasaccharide partially occupies the HS/heparin binding site I. (C) Model (pose-b) showing interactions of pixatimod with the RBD domain of spike protein wherein the sulfated tetrasaccharide partially occupies the HS/heparin binding site III. The RBD is rendered using the Coulombic surface whereas the ligand is shown as a stick. (D) The lipophilic tail of pixatimod in both models wraps around the hydrophobic residues, thereby creating a steric clash with the helix of ACE2 protein. (E) The RBD is shown as a ribbon colored from the N- to C-terminal (blue to red). The residues of RBD (shown as a ball and stick) responsible for binding to ACE2 (shown in a light blue ribbon) are labeled. Pixatimod uses similar residues from the receptor binding motif on the RBD and thereby inhibits binding of ACE2. The Coulombic surface was rendered using UCSF Chimera coloring defaults: ε = 4r, thresholds ±10 kcal/mol. Red corresponds to negative charges, white to neutral, and blue to positive charges. Hydrogens are not shown for clarity.