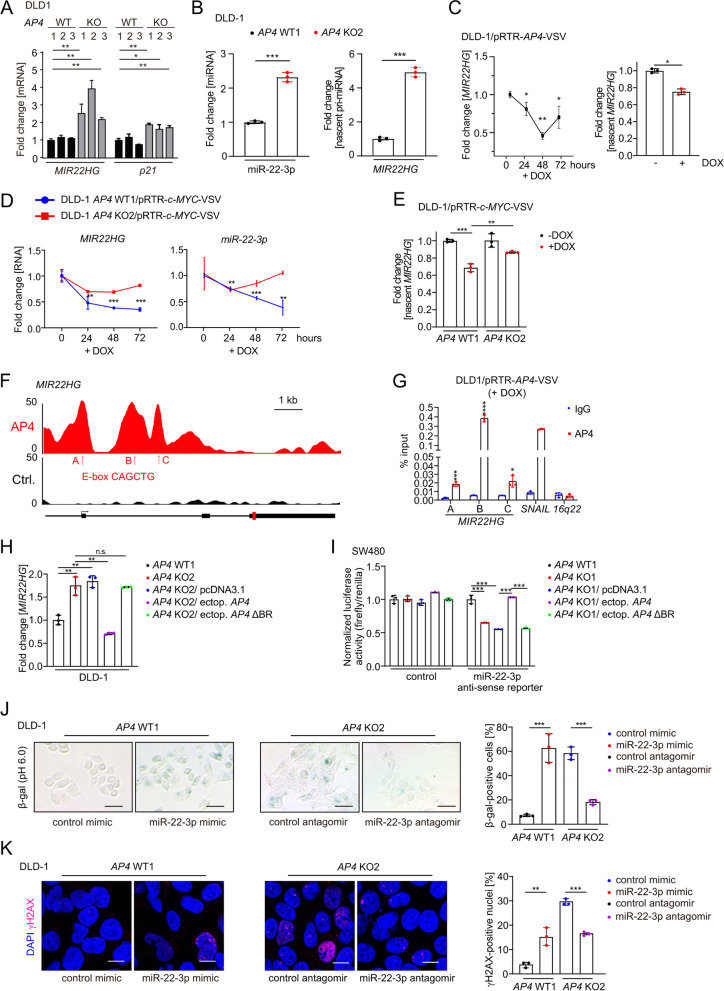
Fig. 2.
AP4 directly represses MIR22HG. A qPCR analysis of MIR22HG and p21 in the indicated clones of DLD-1 cells. B qPCR analysis of miR-22-3p expression (left panel) and nascent MIR22HG mRNA (right panel). C qPCR analysis of MIR22HG expression after AP4 induction by DOX for the indicated duration (left panel) and nascent MIR22HG 48 h after AP4 induction by DOX (right panel). D qPCR analysis of MIR22HG (left panel) and miR-22-3p (right panel) after c-MYC induction by DOX for indicated durations. E qPCR analysis of nascent MIR22HG 48 h after c-MYC induction by DOX. F ChIP-Seq enrichment profiles for AP4-VSV associated chromatin were generated with the UCSC genome browser. The gene structure ideogram is shown below the ChIP-seq tracks. G qChIP analysis of AP4 occupancy. Cells were treated with DOX for 48 h. Chromatin was enriched by anti-AP4 or anti-rabbit-IgG antibodies. SNAI1 and 16q22 served as positive and negative controls, respectively. H qPCR analysis of MIR22HG in the indicated cells 48 h after transfections. I miR-22-3p anti-sense reporter dual-luciferase analysis 48 h after transfection. J β-gal detection at pH 6 48 h after transfection. Quantification of 3 fields with 120 cells in total. Scale bars: 50 μm. K Detection of γH2AX foci 48 h after transfection. Quantification of 3 fields with 120 cells total. Scale bars: 20 μm. In panel A-E and G-K the mean + SD (n = 3) is provided with *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001