Abstract
Simple Summary
Heat stress is a major factor in causing substantial losses to the livestock industry, due to its negative effects on milk production, fertility, health and welfare. As the second largest diary producing species, buffalo face challenges from heat stress. To explore the nutritional metabolic mechanism of rumen bacteria in buffalo under hot enviroments, this study used 16S rDNA technology and metabolome analysis to identify heat stress-related bacteria, metabolites and their pathways. Our findings showed that heat stress increased respiratory rate and skin temperature, and decreased rumen volatile fatty acids. The structures of rumen bacterial communities at different levels significantly changed and a total of 32 metabolites mainly involved in gluconeogenesis were altered due to heat stress. Overall, this study improves our understanding of ruminal bacterial and metabolic alterations of buffalo exposed to heat stress.
Abstract
Exposure to the stress (HS) negatively affects physiology, performance, reproduction and welfare of buffalo. However, the mechanisms by which HS negatively affects rumen bacteria and its associated metabolism in buffalo are not well known yet. This study aimed to gain insight into the adaption of bacteria and the complexity of the metabolome in the rumen of six buffalo during HS using 16S rDNA and gas chromatography metabolomics analyses. HS increased respiratory rate (p < 0.05) and skin temperature (p < 0.01), and it decreased the content of acetic acid (p < 0.05) and butyric acid (p < 0.05) in the rumen. Omics sequencing revealed that the relative abundances of Lachnospirales, Lachnospiraceae, Lachnospiraceae_NK3A20_group and Clostridia_UCG-014 were significantly (p < 0.01) higher under HS than non-heat stress conditions. Several bacteria at different levels, such as Lactobacillales, Streptococcus, Leuconostocaceae and Leissella, were significantly (p < 0.05) more abundant in the rumen of the non-heat stress than HS condition. Thirty-two significantly different metabolites closely related to HS were identified (p < 0.05). Metabolic pathway analysis revealed four key pathways: D-Alanine metabolism; Lysine degradation, Tropane; piperidine and pyridine alkaloid biosynthesis; and Galactose metabolism. In summary, HS may negatively affected rumen fermentation efficiency and changed the composition of rumen community and metabolic function.
Keywords: buffalo, heat stress, rumen fermentation, high-throughput sequencing, metabolomic
1. Introduction
Buffalo, as the second largest producers of meat and milk, are mainly distributed in tropical and subtropical regions of Asia [1]. Buffalo have a higher feed conversion efficiency and higher nutritive value of milk compared to dairy cows [2]. Buffalo present dark skin and sparse hair phenotypes, making the animals highly susceptible to absorb large amounts of solar radiation [3]. In addition, the sweat glands of buffalo are underdeveloped [4]. For this reason, buffalo have a poor heat dissipation capacity that makes them highly restricted to habitat requirements and considerably susceptible to global warming [5]. With the increasing impacts of the climate change, the issue of heat stress (HS) is likely to become worse for buffalo. The temperature-humidity index (THI) has been popularly used to indicate degree of HS. Heat stress in buffalo begin at THI 72, beyond which buffalo are unable to maintain heat balance, causing low dry matter intake (DMI) and high respiratory rate [6]. When the THI exceeds 75, animal productivity, fertility and health have been strongly affected, which subsequently cause economic losses in the dairy industry [7,8].
Most modern farms adopt measures including physical cooling and nutritional regulation to alleviate the impacts of HS on buffalo. However, these strategies may not eliminate completely the negative effects of HS due to animal-related factors including genetics, hair coat and metabolic heat production. With the development of modern molecular genetics technology, genomics techniques have attracted the vision of researchers. Metabolomics has been successfully applied for filtering out biomarkers for milk quality, energy metabolism, and rumen health in dairy cows [9,10]. Previous reports have applied macrobiotics techniques and found that the rumen microbial diversity of cows changes under the influence of HS [11]. Due to the high metabolic heat production with rumen fermentation, buffalo are more susceptible to the negative effects of HS compared with other farm animals [12]. As bioreactors, the rumen has highly dense and diverse microbial populations, which play an important role in buffalo health and metabolism [13]. The knowledge of rumen bacteria and metabolome could assist to better understand the effect of hot environment on animal productivity, physiology and immune activity. In addition, the identification of biomarkers may provide valuable tools to diagnose the incidence of HS in buffalo. Therefore, the objective of this study was to compare the changes in physiology, rumen microbes and their metabolites in buffalo under both HS and non-heat stress (NHS) conditions, so as to further understand how buffalo differentially respond to HS that ultimately could assist in formulating effective strategies to reduce the adverse effects of HS and provide a theoretical basis for improving milk production performance using means such as feeding management.
2. Materials and Method
2.1. Animal Management
Buffalo in this experiment were primarily selected based on days in milk (142 ± 11.1), and finally, six healthy second-parity buffalo of similar body weight (647 ± 24.48 kg) and age (49 ± 1.48 months) were selected. The experiment was conducted at Hubei Prime Cattle Husbandry Co., Ltd. (Jingmen, China) during June and August of 2021 as NHS and HS condition for seven days, respectively. The same management system was applied along the whole experimental period. All the animals were housed in individual pens with free access to water. Total mixed rations (TMR) were fed twice daily and were offered to ensure 10% residual feed. Feed intake was calculated for each cow during the trial. Samples of feed stuffs and TMR were collected on days 1, 4, 7 for each cow for NHS and HS period. Then, feed ingredients and composition were determined according to Association of Official Analytical Chemists (AOAC) standards in the laboratory [14]. Detailed composition and nutrient content of the diets are presented in Table 1.
Table 1.
Feed ingredients and composition of the diets fed to experimental buffalo (n = 6).
| Item | Content |
|---|---|
| Ingredient (%, DM basis) | |
| Silage corn | 30 |
| Rice straw | 45 |
| Peanut vine | 10 |
| Corn | 7 |
| Rice bran | 2.5 |
| Soybean meal | 3 |
| Wheat bran | 2.5 |
| Mineral lick 1 | |
| Nutrient composition | |
| DM, % | 67.50 |
| NDF, % of DM | 32.74 |
| ADF, % of DM | 20.05 |
| CP, % of DM | 11.60 |
| Ca, % of DM | 0.70 |
| P, % of DM | 0.50 |
| GE, Mcal/kg of DM | 7.58 |
Note: provided by Hubei Prime Cattle Husbandry Co., Ltd., Jingmen, China. 1. The ingredients of licking bricks are urea, Nacl, molasses, minerals and multivitamins (Cangzhou Haili brick Co., Ltd., Cangzhou, China). Abbreviations: DM = dry matter; NDF = neutral detergent fiber; ADF = acid detergent fiber; CP = crude protein; GE = gross energy.
2.2. Sample Collection and Measurement
Two distinct time points, as HS and NHS conditions, were determined based on THI. The temperature and humidity were continuously recorded every hour during the trial for calculating THI, which was calculated by using the following formula: THI = (1.8 × Tdb + 32) − [(0.55 − 0.0055 × RH) × (1.8 × Tdb − 26.8)], where Tdb is the dry-bulb temperature (°C), and RH is the relative humidity (%) [4]. Simultaneously, physiological parameters including respiratory rate, rectal temperature and skin temperature were measured three times daily (8:00 a.m., 12:00 p.m. and 17:00 p.m.) throughout the experimental period according to the method described by [8]. Buffalo were milked twice per day, and milk yield was recorded at each milking for each cow. Milk samples were collected during evening milking at day 7 and analyzed for the individual cow for fat, protein, lactose, milk urea nitrogen and SCC at the laboratory of Hubei Dairy Herd Improvement Center. Blood samples from the jugular vein of each cow were taken at 12:00 p.m. on the last day of the experimental period. A 10 mL blood collection tube was used for plasma separation. Then, blood samples were centrifuged at 3500× g for 10 min and stored at −20 °C until performing the analyses of heat shock protein 70 (HSP70) and heat shock protein 90 (HSP90). The serum concentrations of HSP70 and HSP90 were determined by enzyme immunoassay-ELISA commercial reagent kits (Shanghai Enzyme-linked Biotechnology Co., Ltd., Shanghai, China). The respective intra- and inter-assay coefficients of variations were less than 10%.
As previously described by [15], rumen fluid samples were extracted from the rubber end of the rumen tube connected with a syringe posterior on the last day of the NHS and HS periods, prior to morning feeding. The oral stomach tube was inserted to a depth of 200 cm to ensure that it reached the central rumen, thus obtaining representative rumen fluid samples [16]. The first two extractions were discarded for minimizing saliva contamination. The pH of rumen fluid was measured immediately after sample collection, and then stored in sterile tubes and snap-frozen in liquid nitrogen before storage at −80 °C for subsequent DNA extraction and analysis. Total volatile fatty acids (TVFA) were determined by ion chromatography using Dionex ICS-3000 ion chromatograph following the procedure proposed by [17,18]. The concentration of NH3N was measured using visible-light spectrophotometry (Scientific BioMate 3s, Thermo, Waltham, MA, USA).
2.3. DNA Extraction, 16S rDNA Sequencing and Sequences Analysis
After thawing, microbial genomic DNA was extracted from 1 mL rumen samples using a commercial DNA extraction kit (OMEGA Bio-Tek, Norcross, GA, USA), in accordance with the manufacturer’s instructions [19]. DNA concentration was determined using a spectrophotometer (Nanodrop, Thermo Fisher Scientific Oxoid Ltd., Basingstoke, UK). The amplification of 16S rDNA V3 to V4 regions was conducted using the forward primer 338F (5′-ACTCCTACGGGAGGCAGC-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′) [20]. The 5′ end of the primers in each sample was marked with specific barcodes. The specific PCR products were detected and quantified using the QuantiFluor™-ST blue fluorescence quantitation system (Promega Co., Ltd., San Diego, CA, USA), followed by mixing in equal amounts. The detailed PCR procedure applied was according to that described by [20]. The amplicon library was quantified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and assessed with the Hieff NGSTM Library Quantification Kit for Illumina (Kapa Biosciences, Woburn, MA, USA). After the constructed library was qualified, the Illumina PE250 system was used for sequencing (Shanghai Majorbio Bio-pharm Technology Co., Ltd., Shanghai, China). Raw sequencing reads have been deposited in the NCBI Sequence Read Archive (SRA) under Bio Project PRJNA793724.
Following sequencing, raw sequences were filtered and classified using MOTHUR (v1.30.2). The sequence shorter than 200 bp, or with a homopolymer longer than 8 bp and the raw reads containing ambiguous bases were removed to ensure high accuracy for the subsequent analysis. Non-repetitive sequences (excluding single sequences) were clustered for operational taxonomic unit (OTU) following 97% similarity (UPARSE, version 7.0.1090), based on a representative sequence of the resulting OTU. Analyses of bacterial abundances and diversity index, and the representative sequence of each OTU were aligned against the Silva database (Release138 http://www.arb-silva.de, accessed on 15 October 2021). The overall microbiotas shaped by the two different temperature conditions throughout the experimental period were measured using principal component analysis (PCA) and principal co-ordinates analysis (PCoA) by means of QIIME software (v1.9.1)
2.4. Metabolomics Profiling for Rumen Fluid and Data Analysis
Rumen samples were prepared to be pretreated, extracted and derivatized according to the method described by [21]. Quality control samples were prepared to evaluate the stability of the analytical system. Furthermore, the quality control samples with relative standard deviation ≥ 30% were deleted. In brief, a 100 μL biofluid sample was mixed with 300 μL methanol, followed by adding 50 μL L-2-chlorophenylalanine, and then the mixture was vortexed. Subsequently, all sample mixtures were kept at −20 °C for 20 min and centrifuged for 15 min (14,000× g, 4 °C). The extracts were dried using a vacuum concentrator, then a volume of 80 μL of o-methyl hydroxylamine hydrochloride was added, and finally the solution was vortexed again and maintained for further analysis.
The gas chromatography metabolomics (GC/MS) data analysis was conducted by integrating each resolved chromatogram peak. The metabolite annotation and data pretreatment were conducted on the free online platform of Majorbio Cloud Platform (www.majorbio.com, accessed on 15 October 2021). After raw data collection, orthogonal projections to latent structures discriminant analysis (OPLS-DA) were carried out by ropls (R packages, Version 1.6.2). The metabolites were identified and confirmed by using Kyoto Encyclopedia of Genes and Genomes (KEGG), whereas MetaboAnalyst 4.0 was performed to obtain the relevant pathways [22]. Furthermore, the significantly differential metabolites were screened using variable importance in projection (VIP) scores (VIP >1) from the OPLS-DA, p-values (p < 0.05) and the fold change (FC) (log2 FC > 1 or <−1).
2.5. Statistical Analysis
The correlations between altered rumen bacteria and various heat stress response indices, including the physiological parameters, animal performance and rumen fermentation were assessed by Pearson correlation analyses, where p-value of less than 0.05 was considered statistically significant. Physiological parameters, blood parameters, animal performance and rumen fermentation were tested for normality using Shapiro–Wilk’s test, and the data conformed to a normal distribution. Since SCC is a skewed distribution, convert it to somatic cell score (SCS) according to the formula for the analysis: SCS = Log2 (SCC/100) + 3 [23]. Then, the data were statistically analyzed by the linear mixed model. The statistical model included the fixed effects of factors (the HS and NHS conditions) and covariates (DIM and DMI); the random effect of the animal. Pearson correlation analyses, normal distribution test and linear mixed model were performed using SPSS 25.0 software (IBM Inc., Chicago, IL, USA). Significant differences were declared at p < 0.05, whereas p < 0.01 was considered highly significant. The results of these analyses were expressed as means and standard error of the means (SEM).
3. Results
3.1. Physiological Parameters, Animal Performance and Rumen Fermentation
Measures of THI recorded in June as NHS condition (THI = 70.31), and in August, as HS condition (THI = 80.77) indicated that the buffalo individuals were exposed to HS during August. As shown in Table 2, respiratory rate (p < 0.01) and skin temperature (p < 0.05) were increased in the HS condition compared with the NHS condition. Regarding rumen fermentation, values of acetic acid (p < 0.05), butyric acid (p < 0.05), TVFA (p < 0.05) and pH (p < 0.05) were lower in the HS condition as compared with the NHS condition.
Table 2.
Differential response in physiological parameters, blood parameters, animal performance, and rumen fermentation for buffalo under non-heat stress (NHS; n = 6) and heat stress (HS; n = 6) conditions.
| Index | NHS | HS | SEM | p-Value |
|---|---|---|---|---|
| Physiological parameters | ||||
| Respiratory rate (bpm) | 46.39 | 66.27 | 6.74 | 0.005 |
| Rectal temperature (°C) | 38.74 | 38.80 | 0.14 | 0.831 |
| Skin temperature (°C) | 35.66 | 38.22 | 0.71 | 0.010 |
| Blood parameters | ||||
| HSP70 (ng/mL) | 395.91 | 568.79 | 117.77 | 0.126 |
| HSP90 (ng/mL) | 236.84 | 376.33 | 113.90 | 0.525 |
| Animal performance | ||||
| DMI (kg/d) | 13.03 | 11.57 | 1.29 | 0.456 |
| Milk yield (kg/d) | 4.10 | 2.93 | 1.72 | 0.480 |
| Fat (%) | 8.66 | 7.71 | 1.07 | 0.415 |
| Protein (%) | 4.76 | 4.47 | 0.49 | 0.064 |
| Milk urea nitrogen (mg/dL) | 14.64 | 20.48 | 3.42 | 0.060 |
| SCS | 3.68 | 3.88 | 1.26 | 0.581 |
| Rumen fermentation | ||||
| pH | 6.85 | 6.63 | 0.15 | 0.031 |
| Acetic acid (mmol/L) | 44.72 | 30.07 | 6.38 | 0.030 |
| Propionic acid (mmol/L) | 13.29 | 11.22 | 1.86 | 0.467 |
| Butyric acid (mmol/L) | 9.38 | 7.98 | 0.81 | 0.016 |
| TVFA (mmol/L) | 67.39 | 49.27 | 8.35 | 0.038 |
| A:P | 3.36 | 2.76 | 0.32 | 0.089 |
| NH3N (mg/dL) | 5.33 | 10.76 | 2.49 | 0.059 |
Abbreviations: bpm = breath per minute; HSP = heat stress protein; DMI = dry matter intake; SCS = somatic cell score; TVFA = total volatile fatty acid; A:P = acetic acid to propionic acid ratio. Note: Physiological parameters were measured three times daily (08:00 a.m., 12:00 p.m. and 17:00 p.m.); DMI and milk yield were recorded twice daily; milk samples were collected during evening milking at day 7 and analyzed for milk composition; blood and rumen fluid samples were collected on the last day of the experimental period, prior to morning feeding.
3.2. Rumen Bacteria Diversity and Composition
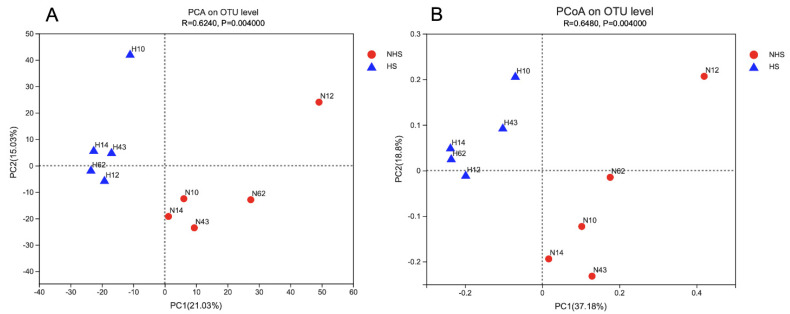
High throughput sequencing of the 16S rDNA gene revealed that a total of 677,955 raw reads were obtained, whereas 39,870 OTU were obtained by performing OTU clustering on nonrepetitive sequences according to 97% similarity. The detailed sequence information is provided in Table S1. As shown in Table 3, Good’s coverage values for the two periods were greater than 0.99, indicating good sequencing depth for analysis of the rumen microbiota. Microbial community richness and diversity, as represented by Sobs, ACE and Shannon diversity indices of the microbial communities in the rumen of buffalo, showed no significant difference between NHS and HS conditions. Additionally, PCA and PCoA based on Bray–Curtis distance were performed to compare the composition of bacterial communities under both NHS and HS conditions revealing that the bacterial communities were obviously separated by HS (Figure 1).
Table 3.
Effects of HS on richness and diversity of rumen bacteria in buffalo (n = 6).
| Index | NHS | HS | SEM | p-Value |
|---|---|---|---|---|
| Sobs | 1487.40 | 1581.80 | 89.51 | 0.351 |
| ACE | 1738.79 | 1842.35 | 139.09 | 0.498 |
| Shannon | 5.43 | 5.68 | 0.15 | 0.170 |
| Coverage | 0.99 | 0.99 | 0.00 | 0.835 |
Abbreviations: NHS = non−heat stress; HS = heat stress; ACE = abundance−based coverage estimator.
Figure 1.
Beta diversity of ruminal microbiota using principal component analysis (A) and principal co-ordinates analysis (B). NHS, non-heat stress condition; HS, heat stress condition.
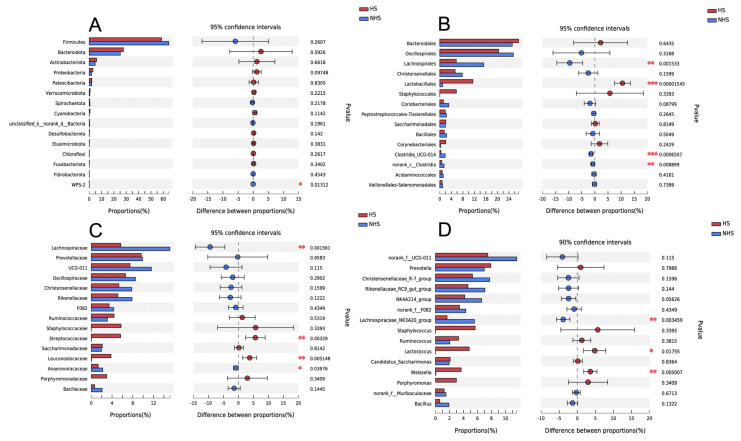
Ruminal bacterial community composition and structure were analyzed at different taxonomical levels. At the phylum level (Figure 2A), most sequences were assigned to Firmicutes (58.56–64.41) and Bacteroidota (25.14–27.73). Four predominant taxa at order level including Lachnospirales, Lactobacillales, Clostridia_UCG-014, and norank_c__Clostridia were significantly shifted (p < 0.01) during the period of HS (Figure 2B). Significant shifts were also detected at family level (Figure 2C). The relative abundances of Lachnospiraceae and Anaerovoracaceae were significantly (p < 0.05) higher in HS condition, whereas Streptococcaceae and Leuconostocaceae were significantly (p < 0.01) lower in HS condition. In addition, HS resulted in a decrease (p < 0.05) in genus level in the relative abundances of Lactococcus and Weissella, and a significant (p < 0.01) elevation in the relative abundances of Lachnospiraceae_NK3A20_group (Figure 2D).
Figure 2.
Rumen bacteria affected by heat stress at different taxonomical levels. (A) phylum (B) order level (C) family level (D) genus level. * represents p < 0.05, ** represents p < 0.01, *** represents p < 0.001. NHS, non-heat stress condition; HS, heat stress condition.
3.3. Identification of Different Metabolites and Metabolic Pathways
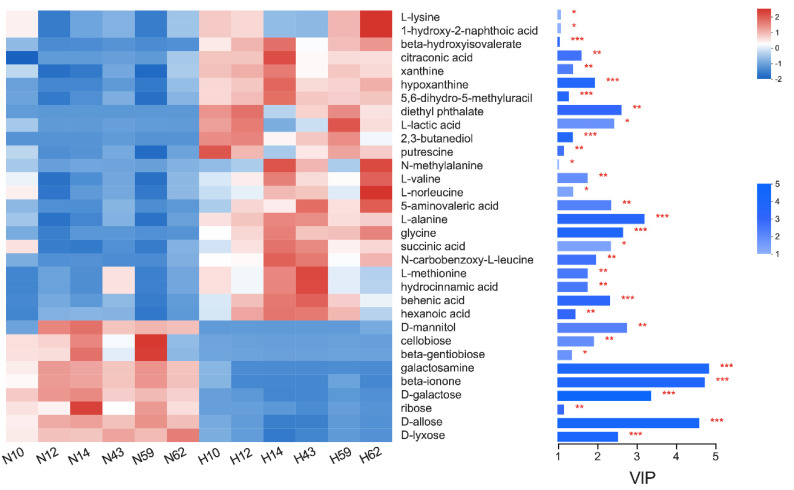
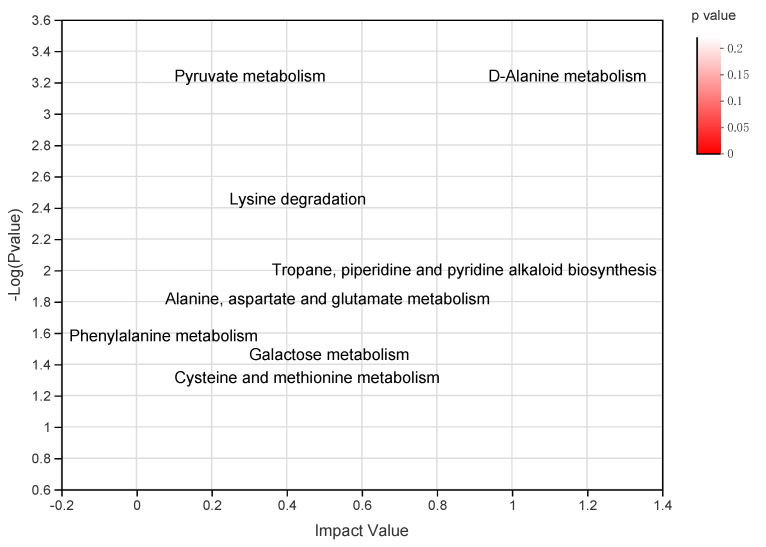
The OPLS-DA analysis of GC-MS metabolic profiles of rumen fluid showed a separation between the NHS and HS (Figure S1). For the rumen metabolome, we analyzed the 197 compounds identified in the present study. After t-test and VIP filtering for the relative concentrations of rumen metabolites, the comparison analysis revealed that the concentrations of 9 metabolites were significantly (p < 0.05, VIP > 1) higher in the rumen of animals under HS compared to NHS conditions, whereas 23 metabolites were significantly (p < 0.05, VIP > 1) lower in the rumen of buffalo assigned to HS compared to NHS condition (Figure 3). The metabolome view map based on 32 differential metabolites revealed the enrichment of 8 pathways (p < 0.05). However, only 4 of them were characterized as significantly relevant pathways which had a pathway impact value higher than 0.1, involved in D-Alanine metabolism, Lysine degradation, Tropane, piperidine and pyridine alkaloid biosynthesis, and Galactose metabolism (Figure 4).
Figure 3.
Differential metabolites in rumen fluid of buffalo under non-heat stress and heat stress conditions. The heatmap is the metabolite expression and the right side is the metabolite variable importance in projection (VIP) bar graph. The color of the bar indicates the significance of the difference between the two groups of metabolites, * represents p < 0.05, ** represents p < 0.01, *** represents p < 0.001. N, non-heat stress condition; H, heat stress condition.
Figure 4.
Key differential metabolic pathways characterize in the rumen fluid of buffalo under non-heat stress and heat stress conditions. The x-axis represents the pathway impact, and y-axis represents the pathway enrichment.
3.4. Correlation between Rumen Bacteria, Metabolites and Fermentation Parameters
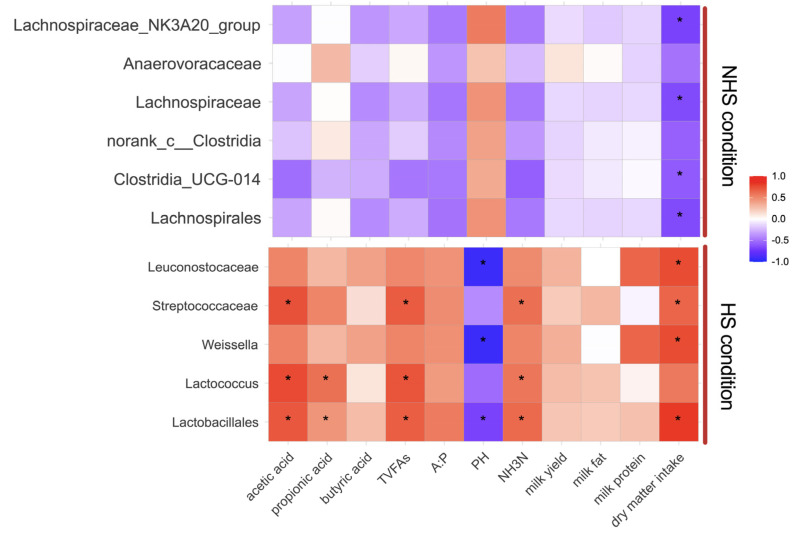
To explore the relationships between rumen fermentation, physiological and blood parameters, animal performance and rumen bacteria, Pearson correlation coefficients were calculated for the HS and NHS conditions (Figure 5). Significantly negative correlations were found between Lachnospirales, Lachnospiraceae_NK3A20_group, Lachnospiraceae and Clostridia_UCG-014 with dry matter intake (r < 0.5, p < 0.05). We also detected that Lactobacillales, Streptococcaceae and Weissella were positively correlated with NH3N, acetic acid and VFA (r < 0.5, p < 0.05).
Figure 5.
Effect of heat stress on correlation analysis between rumen bacteria and heat stress response indices including physiological and blood parameters, animal performance and rumen fermentation were assessed by Pearson correlation analyses. NHS, non-heat stress condition; HS, heat stress condition. Red indicates a positive correlation; the blue indicates a negative correlation; asterisks denote significant differences at p < 0.05.
4. Discussion
Heat stress is a severe challenge for buffalo. Previous studies have found that farm animals display differential HS responses at the ambient temperature [24,25,26]. In the present study, the significant differences between NHS and HS conditions in terms of their effects on respiratory rate and skin temperature reveal buffalo sensitivity to hot environments. Due to fewer sweat glands, buffalo lose 88% of the internally produced heat, whereas the remaining body heat dissipates from the skin surface by sweating [6]. Previous studies in dairy cows have found a reduction in milk yield by 30–40% due to HS, and this decrease in milk production can be partly attributed to reduced feed intake [27,28]. In addition, an increase in rectal temperature by 1 °C under HS conditions was sufficient to reduce the performance of dairy cows [29]. However, the negative effect of heat stress on milk yield and feed intake in dairy cows is not reflected on buffalo individuals in our study. We speculate that this discrepancy may be attributable to the fact that higher producing dairy cows with greater growth and metabolic activity compared to buffalo, the body heat load is increased, thus amplifying heat strain in dairy cows [30]. The difference in reaction to HS may be also related to variation in heat tolerance ability among between different breeds and individuals. Further investigations trials are needed to expand the sample size and compare the response to heat stress among individuals with different heat tolerance in the same breed. HS altered buffalo rumen fermentation in the present study, including a decrease in acetic and butyric acid, which are precursors of a diverse range of compounds in the body especially fatty acids and total cholesterol [31]. Previous studies have documented that differential basic metabolism of VFA in rumen caused variation in milk quality, as acetic acid affects milk fat synthesis, and propionic acid is involved in the biosynthesis of milk protein and lactose [32,33].
Rumen fermentation has a dynamic response to alterations in the rumen microbiome [34]. The 16S rDNA gene sequencing analysis (V3–V4) used in this study only focused on bacteria, which are the most predominate and diverse among the ruminal microbiota (archaea, fungi, protozoa and bacteria) [35]. The rumen is a major habitat for numerous species of microbes, where fluctuations in the number and composition of rumen bacteria reflect changes in nutrients and rumen physiological environments [36]. In the current study, HS had no significant influence on the alpha diversity of the rumen bacteria. On the contrary, the analysis of β-diversity showed noticeable changes in the bacterial community composition owing to HS. We found that the bacteria of the rumen were mainly composed of Firmicutes and Bacteroidota (the relative abundance > 5%), which is in agreement with previous studies in buffalo [37]. Regarding the relative abundances of major order and family, we found that the relative abundances of Lactobacillales, Streptococcus and Leuconostocaceae were higher under the HS compared with NHS condition, and lactic acid is the main end-product of their characteristic heterofermentative carbohydrate metabolism [38]. Of note, the absorption rate of lactic acid from the rumen epithelium is slow, thereby reducing rumen pH [31]. Corresponding to this, the present study also showed that HS significantly decreased pH of rumen, which was negatively correlated with the relative abundances of Lactobacillales and Leuconostocaceae. In addition, Lactobacillales was positively correlated with acetic acid and VFA in our study. Taken together, Lactobacillales and Leuconostocaceae may play a major role in the decline of rumen pH due to HS, and further affect the living environment of other bacterial types. Leuconostocaceae species are used for the production of fermented milk, butter, and cheese together with Lactococcus, Streptococcus and Lactobacillus species [39]. However, what roles the bacterium plays in determining milk quantity and quality under HS and their underlying mechanisms are still unclear.
Acetic and propionic acids are responsible for the formation of fatty acids and cholesterol, which are related to milk fat content, and failure to ensure glucose production when propionic acid is insufficient leads to insufficient energy supply [40,41]. As reported earlier, acetic-acid-producing bacteria and propionic-acid-producing bacteria such as Christensenellaceae_R-7_group, Rikenellaceae_RC9_gut_group and Ruminococcus may contribute to HS responses in cows by increasing heat production in the rumen metabolism [34,42]. However, these genera of bacteria did not differ significantly between the two periods investigated in the present study. It could be concluded that increasing the numbers of lactate-producing bacteria resulted in extra production of lactate and a reduction in the rumen pH under HS condition, which is consistent with the findings in dairy cows [31]. Additionally, Weissella, a genus of Gram-positive bacteria under Leuconostocaceae family, can synthesize various bioactive compounds resulting in a better bioavailability, as well as producing glucan [43]. The microbial composition of the rumen is influenced by various factors such as breed, feeding regimen, age, genetics and sex [44]. The complex relationships between the dynamic changes in rumen microbiota induced by HS, on one side, and feed utilization and production efficiency, on the other side, in domestic buffalo require further investigation. Since the effect of HS on bacteria, archaea, fungi, and protozoa of rumen microbiota may vary differently [45], the microbial diversity and the host-microbiota interaction due to HS also requires further research.
Metabolomics play important roles in understanding physiological and biochemical status of animals [46]. In this study, GC-MS was used to detect the changes in ruminal metabolites from buffalo under NHS and HS conditions. Alanine is the main precursor of glucose production via gluconeogenesis [47]. It has been reported that accumulation of alanine as a major amino acid is increased under HS conditions [48,49]. Our finding is strongly consistent with L-alanine significant increase, indicating the synthesis of glucose by buffalo during HS period and that the cellular energy matrix is insufficient. In ruminants, the gluconeogenic pathway is active and the rumen microorganisms break down cellulose to generate VFA, which are converted to succinyl CoA to participate in the gluconeogenic pathway for glucose synthesis [50,51]. In addition, lactic, citric and succinic acids with greater content in the HS condition can be converted to oxaloacetic acid and then enter the gluconeogenic pathway [52]. Although the high content of glucogenic amino acids (lysine, valine, alanine, glycine and L-methionine) are precursors of milk protein in the HS condition, they are preferentially used to generate tricarboxylic acid intermediates (citric acid, succinic acid, etc.) to participate in the gluconeogenic pathway and amino acid metabolism under HS conditions to maintain energy balance [53,54,55]. We hypothesize that these amino acids are capable of alleviating the adverse effects of HS on buffalo, enhance immunity and respond to anti-oxidation.
This study also deals with most influential pathways involved in significantly different metabolites. In the pyruvate metabolism pathway, on the one hand, pyruvate acts as the end product of glycolysis, and is dehydrogenated to lactate; on the other hand, pyruvate is converted into non-essential amino acids by the action of transaminases and participates in the tricarboxylic acid (TCA) cycle through generating acetyl CoA [56], and then produces adenosine triphosphate, thus providing energy required for milk protein synthesis in dairy cattle [9]. The high concentration of lactic acid and non-essential amino acids in the HS condition of the current study supports the concept of energy supply to the buffalo body through the pyruvate metabolic pathway during the HS condition, suggesting that this pathway may contribute to more active redox reactions to produce more metabolic energy in response to HS. Furthermore, HS affects the metabolism of glucose through diverse pathways such as glycolysis, TCA cycle and galactose metabolism. HS also influences the metabolism of glutamic acid that is an important metabolite in the TCA cycle, leading to a significant decrease in intracellular glutathione levels as well as an increase in reactive oxygen species and oxidative stress [52]. The above-mentioned pathways broadly illustrate that amino acids and energy levels can maintain energy balance under the HS conditions and thus play a fundamental role in buffalo against HS.
5. Conclusions
In general, HS impacted the physiological status of buffalo regarding an increase in respiratory rate and skin temperature. In addition, HS may negatively affected rumen fermentation efficiency and altered the bacterial community and metabolomic profiles in the rumen. Data analysis demonstrated that the structures of rumen bacterial communities at different levels significantly changed under the HS condition compared to NHS condition. The results showed that a total of 32 metabolites mainly involved in gluconeogenesis were altered due to HS. These findings provide new insights to better understand the impacts of HS on rumen fermentation and metabolic disorders. Nevertheless, further research is needed to validate the results of the present study.
Acknowledgments
The authors are grateful to Shanghai Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China. for assisting in sequencing and bioinformatics analysis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani12101300/s1, Figure S1: Orthogonal partial least squares discriminant analysis (OPLS-DA) plot (A) and response permutation testing (B) of rumen metabolites in comparisons of the non-heat stress (NHS) and heat stress (HS) conditions; Table S1: Detailed sequence information of rumen samples of buffaloes using high-throughput sequencing
Author Contributions
Conceptualization, S.L.; methodology, K.N., M.Z. and S.L.; formal analysis, Z.W., K.N., H.E.R. and M.Z.; resources, T.G., F.L., L.Y. and T.F.; investigation, K.N. and S.L.; data curation, Z.W.; writing—original draft preparation, Z.W. and H.E.R.; writing—review and editing, H.E.R. and S.L.; supervision, F.L., L.Y. and T.F.; funding acquisition, T.G. and S.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Animal care and experimental procedures were conducted according to the guidelines of the Animal Care and Use Committee of Henan Agricultural University, Henan Agricultural University, Zheng Zhou, China (Approval number: HENAU-2021-025).
Data Availability Statement
All the analyzed datasets in the current study are available from the corresponding author on reasonable request. The sequence data reported in this paper have been deposited in the NCBI database (BioProjec: PRJNA793724).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the China Agriculture Research System of MOF and MARA, China Postdoctoral Science Foundation (2020M672233) and the Key Scientific Research Project of Henan Province Higher Education (22A230005).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Seerapu S.R., Kancharana A.R., Chappidi V.S., Bandi E.R. Effect of microclimate alteration on milk production and composition in Murrah buffaloes. Vet. World. 2015;8:1444–1452. doi: 10.14202/vetworld.2015.1444-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J. The response of the distributions of Asian buffalo breeds in China to climate change over the past 50 years. Livest. Sci. 2015;180:65–77. doi: 10.1016/j.livsci.2015.07.005. [DOI] [Google Scholar]
- 3.Dayal S., Dey A., Pandian S.J., Gupta J.J., Chandran P.C., Ali I. Effect of seasonal variation on physiological parameters in Murrah buffaloes. Indian J. Anim. Sci. 2017;87:965–967. [Google Scholar]
- 4.Liu S., Li J., Li Z., Deng T., Rehman Z.U., Zichao Z., Liguo Y. Effect of season and breed on physiological and blood parameters in buffaloes. J. Dairy Res. 2018;85:181–184. doi: 10.1017/S0022029918000286. [DOI] [PubMed] [Google Scholar]
- 5.Bombade K., Kamboj A., Alhussien M.N., Mohanty A.K., Dang A.K. Diurnal variation of milk somatic and differential leukocyte counts of Murrah buffaloes as influenced by different milk fractions, seasons and parities. Biol. Rhythm. Res. 2018;49:151–163. doi: 10.1080/09291016.2017.1345472. [DOI] [Google Scholar]
- 6.Gu Z.B., Li L., Tang S.K., Liu C.B., Fu X.H., Shi Z., Mao H. Metabolomics Reveals that Crossbred Dairy Buffaloes Are More Thermotolerant than Holstein Cows under Chronic Heat Stress. J. Agric. Food Chem. 2018;66:12889–12897. doi: 10.1021/acs.jafc.8b02862. [DOI] [PubMed] [Google Scholar]
- 7.Kumar A., Mehrotra S., Singh G., Narayanan K., Das G.K., Soni Y.K., Singh M., Mahla A.S., Srivastava N., Verma M.R. Sustained delivery of exogenous melatonin influences biomarkers of oxidative stress and total antioxidant capacity in summer-stressed anestrous water buffalo (Bubalus bubalis) Theriogenology. 2015;83:1402–1407. doi: 10.1016/j.theriogenology.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 8.Amamou H., Beckers Y., Mahouachi M., Hammami H. Thermotolerance indicators related to production and physiological responses to heat stress of holstein cows. J. Therm. Biol. 2019;82:90–98. doi: 10.1016/j.jtherbio.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 9.Wu X.H., Sun H.Z., Xue M.Y., Wang D.M., Guan L.L., Liu J. Serum metabolome profiling revealed potential biomarkers for milk protein yield in dairy cows. J. Proteom. 2018;184:54–61. doi: 10.1016/j.jprot.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Sun H.-Z., Wang D.-M., Wang B., Wang J.-K., Liu H.-Y., Guan L.L., Liu J.-X. Metabolomics of Four Biofluids from Dairy Cows: Potential Biomarkers for Milk Production and Quality. J. Proteome Res. 2015;14:1287–1298. doi: 10.1021/pr501305g. [DOI] [PubMed] [Google Scholar]
- 11.Zhao S., Min L., Zheng N., Wang J. Effect of Heat Stress on Bacterial Composition and Metabolism in the Rumen of Lactating Dairy Cows. Animals. 2019;9:925. doi: 10.3390/ani9110925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J., Ye G., Zhou Y., Liu Y., Zhao L., Liu Y., Chen X., Huang D., Liao S.F., Huang K. Feeding glycerol-enriched yeast culture improves performance, energy status, and heat shock protein gene expression of lactating Holstein cows under heat stress. J. Anim. Sci. 2014;92:2494–2502. doi: 10.2527/jas.2013-7152. [DOI] [PubMed] [Google Scholar]
- 13.Kim D.-H., Kim M.-H., Kim S.-B., Son J.-K., Lee J.-H., Joo S.-S., Gu B.-H., Park T., Park B.-Y., Kim E.-T. Differential Dynamics of the Ruminal Microbiome of Jersey Cows in a Heat Stress Environment. Animals. 2020;10:1127. doi: 10.3390/ani10071127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.AOAC . Official Methods of Analysis of AOAC International. 17th ed. AOAC; Gaithersburg, MD, USA: 2000. [Google Scholar]
- 15.Liang Y.S., Li G.Z., Li X.Y., Lü J.Y., Li F.D., Tang D.F., Li F., Deng Y., Zhang H., Wang Z.L., et al. Growth performance, rumen fermentation, bacteria composition, and gene expressions involved in intracellular pH regulation of rumen epithelium in finishing Hu lambs differing in residual feed intake phenotype. J. Anim. Sci. 2017;95:1727–1738. doi: 10.2527/jas.2016.1134. [DOI] [PubMed] [Google Scholar]
- 16.Shen J.S., Chai Z., Song L.J., Liu J.X., Wu Y.M. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J. Dairy Sci. 2012;95:5978–5984. doi: 10.3168/jds.2012-5499. [DOI] [PubMed] [Google Scholar]
- 17.Li M., Long S., Wang Q., Zhang L., Hu J., Yang J., Cheng Z., Piao X. Mixed organic acids improve nutrients digestibility, volatile fatty acids composition and intestinal microbiota in growing-finishing pigs fed high-fiber diet. Asian-Australas. J. Anim. Sci. 2019;32:856–864. doi: 10.5713/ajas.18.0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seankamsorn A., Cherdthong A., So S., Wanapat M. Influence of chitosan sources on intake, digestibility, rumen fermentation, and milk production in tropical lactating dairy cows. Trop. Anim. Health Prod. 2021;53:241. doi: 10.1007/s11250-021-02697-0. [DOI] [PubMed] [Google Scholar]
- 19.Liu H., Hu L., Han X., Zhao N., Xu T., Ma L., Wang X., Zhang X., Kang S., Zhao X., et al. Tibetan Sheep Adapt to Plant Phenology in Alpine Meadows by Changing Rumen Microbial Community Structure and Function. Front. Microbiol. 2020;11:2547. doi: 10.3389/fmicb.2020.587558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y.Q.Q., Xi Y.M.M., Wang Z.D.D., Zeng H.F.F., Han Z.Y. Combined signature of rumen microbiome and metabolome in dairy cows with different feed intake levels. J. Anim. Sci. 2020;98:skaa070. doi: 10.1093/jas/skaa070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang R.Y., Liu Y.J., Yin Y.Y., Jin W., Mao S.Y., Liu J.H. Response of rumen microbiota, and metabolic profiles of rumen fluid, liver and serum of goats to high-grain diets. Animal. 2019;13:1855–1864. doi: 10.1017/S1751731118003671. [DOI] [PubMed] [Google Scholar]
- 22.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D.S., Xia J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng L., Yuxia C., Zichen W., Zipeng L., Ahmad M.J., Ming L., Tengyun G., Shenhe L. The Effect of Exogenous Melatonin on Milk Somatic Cell Count in Buffalo. Pak. Vet. J. 2021;41:152–155. doi: 10.29261/pakvetj/2020.074. [DOI] [Google Scholar]
- 24.Hammami H., Bormann J., M’hamdi N., Montaldo H.H., Gengler N. Evaluation of heat stress effects on production traits and somatic cell score of Holsteins in a temperate environment. J. Dairy Sci. 2013;96:1844–1855. doi: 10.3168/jds.2012-5947. [DOI] [PubMed] [Google Scholar]
- 25.Lina X., Chaoyuan W., Luyu D., Zhiyuan G., Lu Z., Zhengxiang S., Baoming L., Chuntao J. Heat stress alleviation for dairy cows housed in an open-sided barn by cooling fan and perforated air ducting (PAD) system. Int. J. Agric. Biol. Eng. 2017;10:1–10. doi: 10.25165/j.ijabe.20171006.3135. [DOI] [Google Scholar]
- 26.Becker C.A., Collier R.J., Stone A.E. Invited review: Physiological and behavioral effects of heat stress in dairy cows. J. Dairy Sci. 2020;103:6751–6770. doi: 10.3168/jds.2019-17929. [DOI] [PubMed] [Google Scholar]
- 27.Wheelock J.B., Rhoads R.P., VanBaale M.J., Sanders S.R., Baumgard L.H. Effects of heat stress on energetic metabolism in lactating Holstein cows. J. Dairy Sci. 2010;93:644–655. doi: 10.3168/jds.2009-2295. [DOI] [PubMed] [Google Scholar]
- 28.Gao S.T., Guo J., Quan S.Y., Nan X.M., Fernandez M.S., Baumgard L.H., Bu D.P. The effects of heat stress on protein metabolism in lactating Holstein cows. J. Dairy Sci. 2017;100:5040–5049. doi: 10.3168/jds.2016-11913. [DOI] [PubMed] [Google Scholar]
- 29.Khongdee T., Sripoon S., Vajrabukka C. The effects of high temperature and roof modification on physiological responses of swamp buffalo (Bubalus bubalis) in the tropics. Int. J. Biometeorol. 2013;57:349–354. doi: 10.1007/s00484-012-0557-3. [DOI] [PubMed] [Google Scholar]
- 30.Johnson J.S., Scharf B., Weaber R.L., Eichen P.A., Spiers D.E. Patterns of heat response and adaptation on summer pasture: A comparison of heat-sensitive (Angus) and -tolerant (Romosinuano) cattle. J. Therm. Biol. 2012;37:344–350. doi: 10.1016/j.jtherbio.2011.10.014. [DOI] [Google Scholar]
- 31.Wang X., Li X., Zhao C., Hu P., Chen H., Liu Z., Liu G., Wang Z. Correlation between Composition of the Bacterial Community and Concentration of Volatile Fatty Acids in the Rumen during the Transition Period and Ketosis in Dairy Cows. Appl. Environ. Microbiol. 2012;78:2386–2392. doi: 10.1128/AEM.07545-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ungerfeld E.M. Shifts in metabolic hydrogen sinks in the methanogenesis-inhibited ruminal fermentation: A meta-analysis. Front. Microbiol. 2015;6:37. doi: 10.3389/fmicb.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L., Sun D., Mao S., Zhu W., Liu J. Infusion of sodium butyrate promotes rumen papillae growth and enhances expression of genes related to rumen epithelial VFA uptake and metabolism in neonatal twin lambs. J. Anim. Sci. 2019;97:909–921. doi: 10.1093/jas/sky459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hassan F., Tang Z., Ebeid H.M., Li M., Peng K., Liang X., Yang C. Consequences of herbal mixture supplementation on milk performance, ruminal fermentation, and bacterial diversity in water buffaloes. PeerJ. 2021;9:e11241. doi: 10.7717/peerj.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu K., Zhang Y., Yu Z., Xu Q., Zheng N., Zhao S., Huang G., Wang J. Ruminal microbiota-host interaction and its effect on nutrient metabolism. Anim. Nutr. 2021;7:49–55. doi: 10.1016/j.aninu.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailoni L., Carraro L., Cardin M., Cardazzo B. Active Rumen Bacterial and Protozoal Communities Revealed by RNA-Based Amplicon Sequencing on Dairy Cows Fed Different Diets at Three Physiological Stages. Microorganisms. 2021;9:754. doi: 10.3390/microorganisms9040754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aguilar-Marin S.B., Betancur-Murillo C.L., Isaza G.A., Mesa H., Jovel J. Lower methane emissions were associated with higher abundance of ruminal Prevotella in a cohort of Colombian buffalos. BMC Microbiol. 2020;20:364. doi: 10.1186/s12866-020-02037-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frantzen C.A., Kot W., Pedersen T.B., Ardö Y.M., Broadbent J.R., Neve H., Hansen L.H., Dal Bello F., Østlie H.M., Kleppen H.P., et al. Genomic Characterization of Dairy Associated Leuconostoc Species and Diversity of Leuconostocs in Undefined Mixed Mesophilic Starter Cultures. Front. Microbiol. 2017;8:132. doi: 10.3389/fmicb.2017.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng J., Wittouck S., Salvetti E., Franz C.M., Harris H., Mattarelli P., O’Toole P.W., Pot B., Vandamme P., Walter J., et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020;70:2782–2858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
- 40.Li M., Hassan F.U., Tang Z., Guo Y., Liang X., Peng L., Xie H., Yang C. Physiological, oxidative and metabolic responses of lactating water buffaloes to tropical climate of South China. Vet. Med. Sci. 2021;7:1696–1706. doi: 10.1002/vms3.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boutard M., Cerisy T., Nogue P.Y., Alberti A., Weissenbach J., Salanoubat M., Tolonen A.C. Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass. PLoS Genet. 2014;10:e1004773. doi: 10.1371/journal.pgen.1004773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo H., Zhou G., Tian G., Liu Y., Dong N., Li L., Zhang S., Chai H., Chen Y., Yang Y. Changes in Rumen Microbiota Affect Metabolites, Immune Responses and Antioxidant Enzyme Activities of Sheep under Cold Stimulation. Animals. 2021;11:712. doi: 10.3390/ani11030712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kavitake D., Devi P.B., Shetty P.H. Overviewof exopolysaccharides produced by Weissella genus—A review. Int. J. Biol. Macromol. 2020;164:2964–2973. doi: 10.1016/j.ijbiomac.2020.08.185. [DOI] [PubMed] [Google Scholar]
- 44.Bainbridge M.L., Cersosimo L.M., Wright A.-D.G., Kraft J. Rumen bacterial communities shift across a lactation in Holstein, Jersey and Holstein x Jersey dairy cows and correlate to rumen function, bacterial fatty acid composition and production parameters. FEMS Microbiol. Ecol. 2016;92:fiw059. doi: 10.1093/femsec/fiw059. [DOI] [PubMed] [Google Scholar]
- 45.Park T., Ma L., Ma Y., Zhou X., Bu D., Yu Z. Dietary energy sources and levels shift the multi-kingdom microbiota and functions in the rumen of lactating dairy cows. J. Anim. Sci. Biotechnol. 2020;11:66. doi: 10.1186/s40104-020-00461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue M.-Y., Sun H.-Z., Wu X.-H., Liu J.-X., Guan L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome. 2020;8:64. doi: 10.1186/s40168-020-00819-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yue S., Ding S., Zhou J., Yang C., Hu X., Zhao X., Wang Z., Wang L., Peng Q., Xue B. Metabolomics Approach Explore Diagnostic Biomarkers and Metabolic Changes in Heat-Stressed Dairy Cows. Animals. 2020;10:1741. doi: 10.3390/ani10101741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cowley F.C., Barber D.G., Houlihan A.V., Poppi D.P. Immediate and residual effects of heat stress and restricted intake on milk protein and casein composition and energy metabolism. J. Dairy Sci. 2015;98:2356–2368. doi: 10.3168/jds.2014-8442. [DOI] [PubMed] [Google Scholar]
- 49.Guo J., Gao S., Quan S., Zhang Y., Bu D., Wang J. Blood amino acids profile responding to heat stress in dairy cows. Asian-Australas. J. Anim. Sci. 2018;31:47–53. doi: 10.5713/ajas.16.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eger M., Hussen J., Koy M., Dänicke S., Schuberth H.J., Breves G. Glucose transporter expression differs between bovine monocyte and macrophage subsets and is influenced by milk production. J. Dairy Sci. 2016;99:2276–2287. doi: 10.3168/jds.2015-10435. [DOI] [PubMed] [Google Scholar]
- 51.Meijer A.J. Amino acids as regulators and components of nonproteinogenic pathways. J. Nutr. 2003;133:2057s–2062s. doi: 10.1093/jn/133.6.2057S. [DOI] [PubMed] [Google Scholar]
- 52.Shen X., Wang C., Liang N., Liu Z., Li X., Zhu Z.J., Merriman T.R., Dalbeth N., Terkeltaub R., Li C., et al. Serum Metabolomics Identifies Dysregulated Pathways and Potential Metabolic Biomarkers for Hyperuricemia and Gout. Arthritis Rheumatol. 2021;73:1738–1748. doi: 10.1002/art.41733. [DOI] [PubMed] [Google Scholar]
- 53.Davoudi P., Daneshyar M. Effect of different levels of tryptophane on performance, carcass characteristics and blood parameters of broiler chickens under heat stress condition. J. Vet. Res. 2017;72:157–164. [Google Scholar]
- 54.Zhang S., Zeng X., Ren M., Mao X., Qiao S. Novel metabolic and physiological functions of branched chain amino acids: A review. J. Anim. Sci. Biotechnol. 2017;8:10. doi: 10.1186/s40104-016-0139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian H., Zheng N., Wang W., Cheng J., Li S., Zhang Y., Wang J. Integrated Metabolomics Study of the Milk of Heat-stressed Lactating Dairy Cows. Sci. Rep. 2016;6:24208. doi: 10.1038/srep24208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belenky P., Bogan K.L., Brenner C. NAD(+) metabolism in health and disease. Trends Biochem. Sci. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the analyzed datasets in the current study are available from the corresponding author on reasonable request. The sequence data reported in this paper have been deposited in the NCBI database (BioProjec: PRJNA793724).