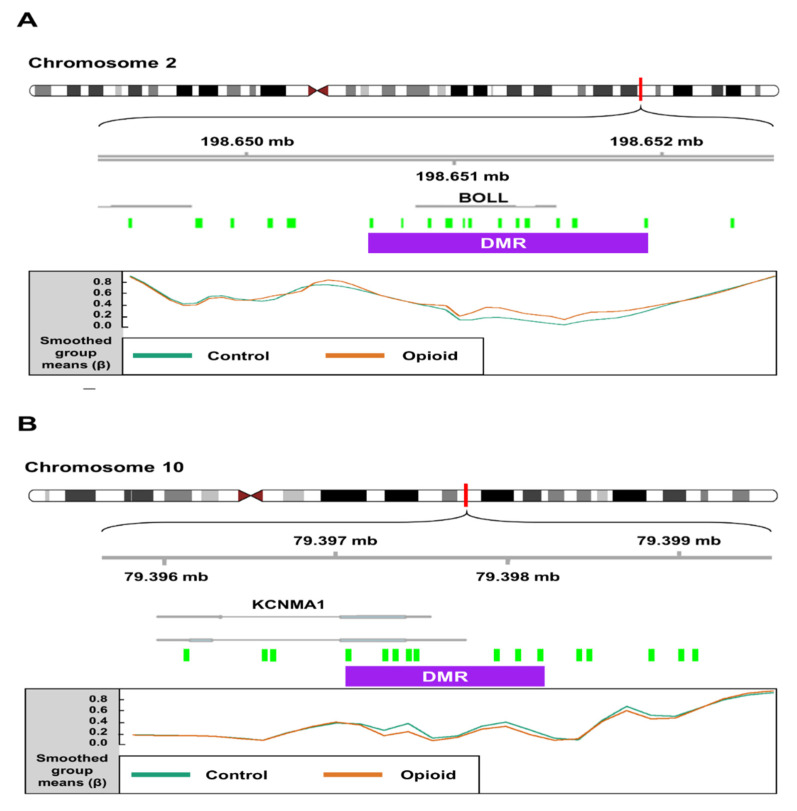
Figure 5.
Schematics depicting differentially methylated regions (DMRs) associated with opioid exposure. (A) A DMR (1,322 bp; represented by purple box) on chromosome 2 spanning 16 CpG probes (green boxes) mapped to the first exon of the Boule Homolog, RNA Binding Protein gene (BOLL). In opioid-exposed samples, methylation levels were 9.02% higher on average at CpG sites within this region relative to control samples (min smoothed FDR = 8.5 × 10−9). (B) A DMR (1120 bp) on chromosome 10 spanning 10 CpG probes mapped to the first exon of the Potassium Calcium-Activated Channel Subfamily M Alpha 1 gene (KCNMA1). Opioid exposed samples had methylation levels that were an average of 11.8% lower than control samples across this region (min smoothed FDR = 2.6 × 10−5). Mean normalized beta values across the DMR are shown for each group (control vs. opioid exposed) at the bottom of each panel.