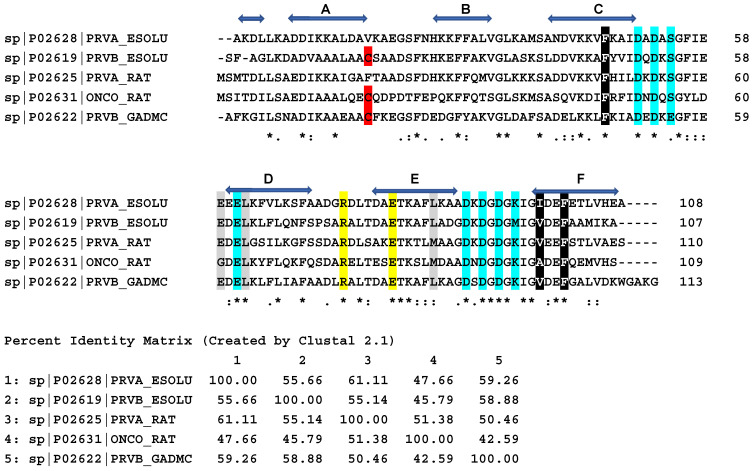
Figure 2.
Amino acid sequences of pike α- and β-PAs, cod β-PA, and rat α- and β-PAs. Horizontal blue arrows show amino acids forming α-helices A through F in the native structure of pike α-PA. The consensus sequence characterizes identical residues as ‘*’, strongly similar residues as ‘:’, and weakly similar residues as ‘.’. Conservative Cys residue in β-PAs is shown in red. Amino acids taking part in calcium coordination are shown in cyan. Conservative Arg and Glu residues forming a salt bridge in the native structure of all PAs are shown in yellow. The data are taken from the UniProt database. Amino acids forming conservative clusters I and II are shown in black and gray, respectively. Percent identity matrix is also shown. Sequences were aligned by Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/; accessed on 23 April 2022).