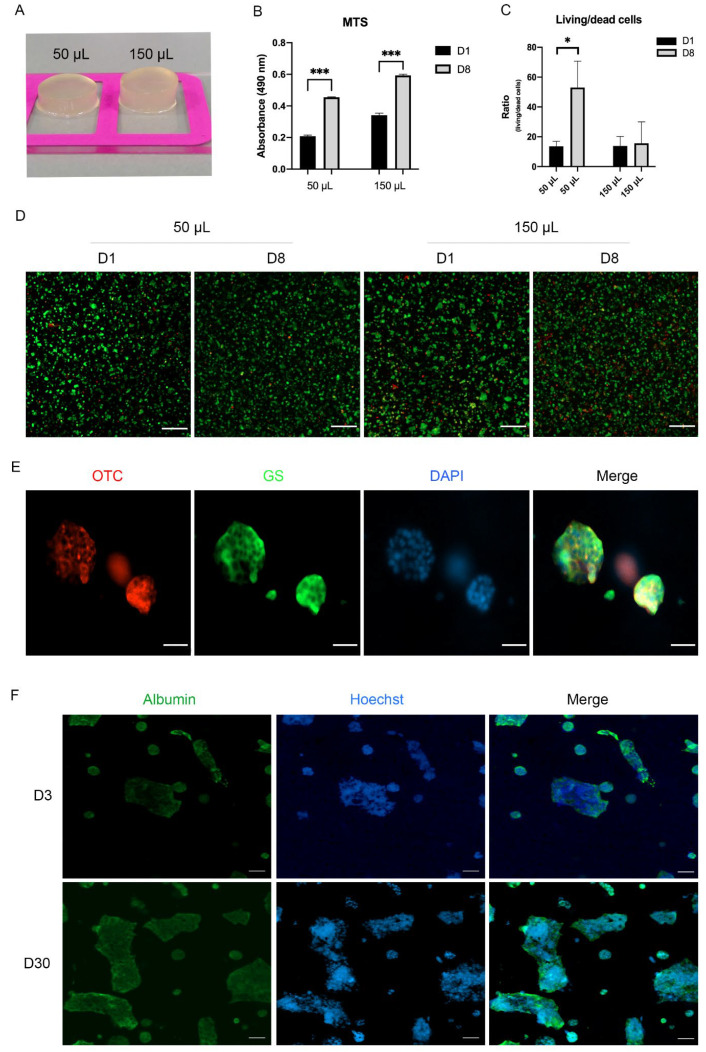
Figure 3.
AML12 cell growth in 3D using GelMA-CMCMA polymer. (A) Picture of AML12 cell growth in 3D using GelMA-CMCMA polymer using two different volumes, cell density 1.5 × 107 cells/mL. (8 mm diameter) (B) Metabolic activity of encapsulated AML12 cells cultured over time. (C) Living-dead cells ratio obtained from 5 random z-Stacks/condition using live/dead assay. (D) Confocal images of live/dead assay of encapsulated AML12 cells. Living cells = green, dead cells = red. Scale bar = 200 µm (E) Confocal images of 3D AML12 cells expressing ornithine transcarbamylase (OTC) and glutamine synthetase (GS). Nuclei stained using DAPI. Scale bar = 50 µm. (F) Confocal images of 3D AML12 cells expressing albumin at indicated timepoints. Nuclei stained using DAPI. Scale bar = 50 µm. The results are expressed as mean values ± SEM and compared using one-way analysis of variance followed by post hoc tests when appropriate. * p < 0.05; *** p < 0.001. MTS: 3-[4,5,dimethylthiazol-2-yl]-5-[3-carboxymethoxy-phenyl]-2-[4-sulfophenyl]-2H-tetrazolium; OTC: ornithine transcarbamylase, GS: glutamine synthetase.