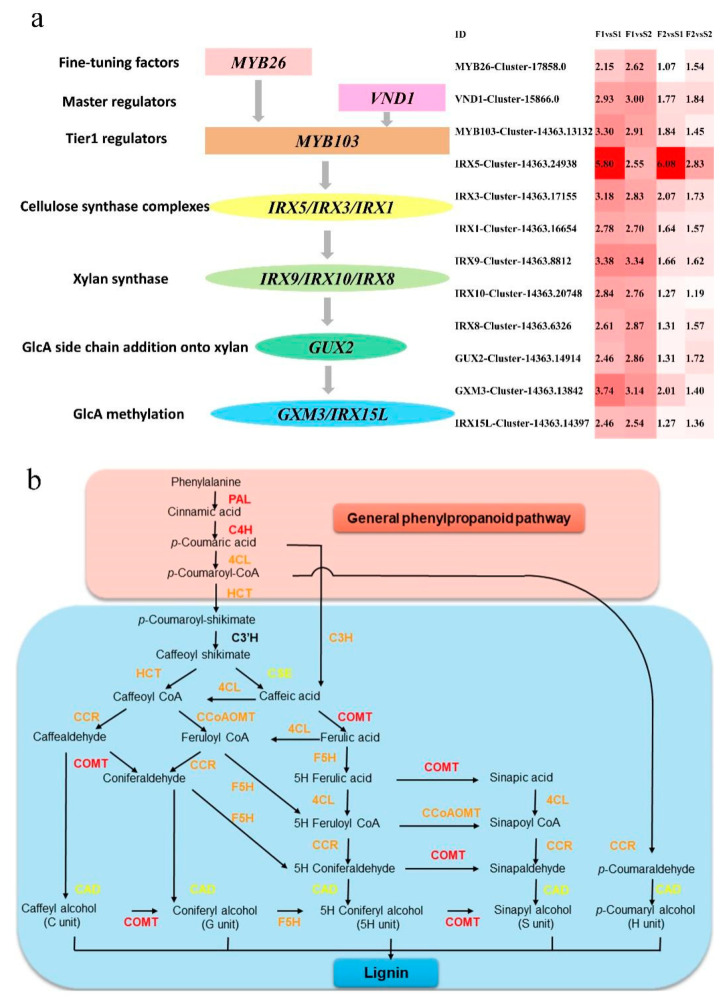
Figure 6.
(a) Illustration of the predicted transcriptional regulating and secondary walls synthesis genes of branch growth in elm. The expression levels (Log2(FC)) of selected differentially expressed genes in fast- and slow-growing cultivars are shown on the right. A red color indicates that the gene is highly expressed in the branch samples. Log2(FC) means Log2() value of fold change for unigenes. (b) The biosynthesis pathway of the general phenylpropanoid and lignin. PAL, phenylalanine ammonia-lyase; TAL, tyrosine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate: CoA ligase; CCR, cinnamoyl-CoA reductase; HCT, hydroxycinnamoyl-CoA shikimate/Quinatehydroxycinnamoyltransferase; CCoAOMT, caffeoyl-CoA O-methyltransferase; F5H, ferulate 5-hydroxylase; CSE, caffeoyl shikimate esterase; COMT, caffeic acid O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase. The color of the proteins indicates the expression of their coding genes in fast and slow growing branches: red indicates up-regulated in the four comparisons (F1vS1, F1vS2, F2vS1, F2vS2), orange indicates up-regulated in three comparisons, yellow indicates up-regulated in two comparisons.