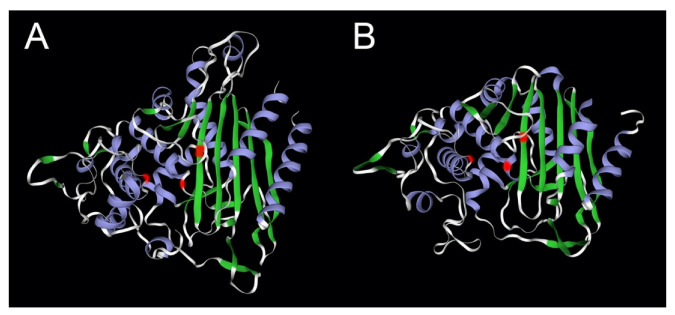
Figure 2.
Comparison of the beta-lactamase fold of LACTB with that of the bacterial PBP-βLs protein flp. Panel (A): LACTB (P83111); panel (B): flp protein of Staphylococcus aureus (Q7A5Q5). The locations of the catalytic site residues in the signature motifs [3] are marked in red, -SISK-, -YST-, and -HTG- for LACTB, and -SNTK-, -YSN-, and -HSG- for the fpl protein. The structure of LACTB was determined by cryo-electron microscopy [2] and that of flp by X-ray crystallography [42]. The image was generated partially using SWISS-MODEL [43].