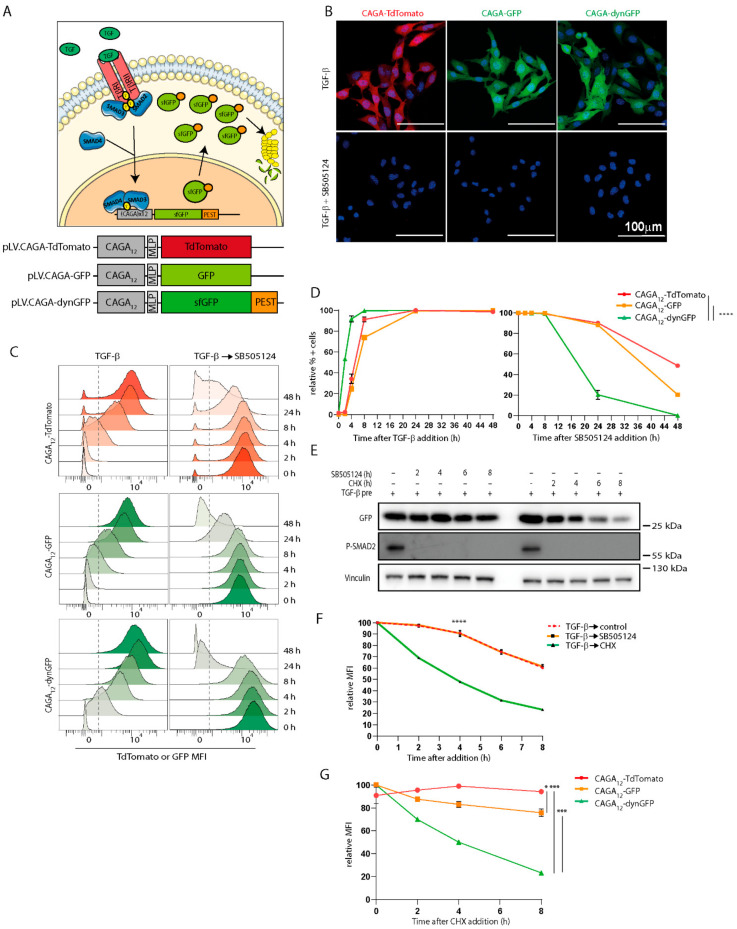
Figure 1.
Generation and validation of the dynamic TGF-β/SMAD3 reporter. (A) Cartoon depicting the TGF-β signaling cascade and activation of the reporter constructs, which are also highlighted below. MLP: major late promoter, sfGFP: superfolder GFP, dynGFP: dynamic GFP. (B) Images of B16F10 cells stably expressing the various TGF-β/SMAD3 reporters treated with TGF-β (1 ng/mL) for 48 h (TdTomato/GFP) or 24 h (dynGFP) or treated with SB505124 (1 mM) together with TGF-β. (C) Flow cytometry of B16F10 cells stably expressing the indicated reporters and treated with TGF-β (1 ng/mL) for 2, 4, 8, 24, and 48 h. SB505124 was added to TGF-β pre-treated (48 h for CAGA12-TdTomato/CAGA12-GFP, 24 h for CAGA12-dynGFP) B16F10 cells. The cut-off for positive cells is shown in the dashed line. (D) Relative percentage of positive cells calculated from (C). Area under the curve (AUC) for TGF-β treatment: CAGA12-TdTomato: 4203 ± 37.01, CAGA12-GFP: 3999 ± 8.40, CAGA12-dynGFP: 4572 ± 13.02. AUC for SB505124 treatment: CAGA12-TdTomato: 3974 ± 34.01, CAGA12-GFP: 3602 ± 40.66, CAGA12-dynGFP: 2001 ± 108.9. One-way ANOVA was performed on AUC values, p < 0.0001. (E,F) The half-life of dynGFP was measured using signal intensity on western blot (E) and relative mean fluorescence intensity (MFI) on flow cytometry (F). Cycloheximide (50 mg/mL) or SB505124 (1 mM) was added after TGF-β pre-treatment (24 h) of CAGA12-dynGFP B16F10 cells. Statistics for all timepoints were determined using two-way ANOVA, shown for timepoint 4 h. (G) The half-life of TdTomato, GFP, and dynGFP were compared by measuring relative MFI on flow cytometry. B16F10 cells were pre-treated with TGF-β (48 h for CAGA12-TdTomato/CAGA12-GFP, 24 h for CAGA12-dynGFP) and treated with cycloheximide (50 mg/mL). Statistics for all timepoints were determined using two-way ANOVA, shown for timepoint 8 h. All experiments were performed in biological triplicates or duplicates (G); error bars represent ± SEM. *** p ≤ 0.001, **** p ≤ 0.0001.